1
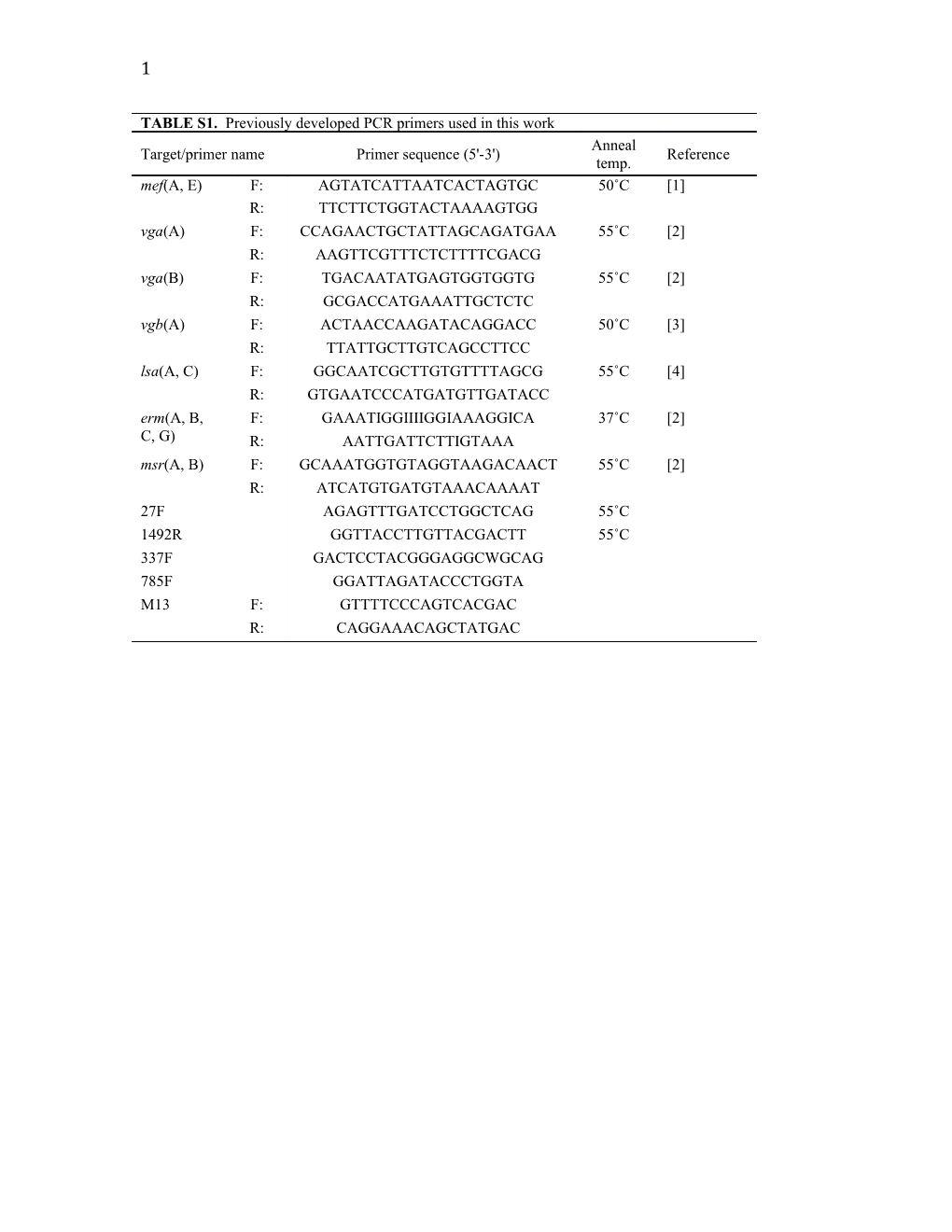
TABLE S1. Previously developed PCR primers used in this work Anneal Target/primer name Primer sequence (5'-3') Reference temp. mef(A, E) F: AGTATCATTAATCACTAGTGC 50˚C [1] R: TTCTTCTGGTACTAAAAGTGG vga(A) F: CCAGAACTGCTATTAGCAGATGAA 55˚C [2] R: AAGTTCGTTTCTCTTTTCGACG vga(B) F: TGACAATATGAGTGGTGGTG 55˚C [2] R: GCGACCATGAAATTGCTCTC vgb(A) F: ACTAACCAAGATACAGGACC 50˚C [3] R: TTATTGCTTGTCAGCCTTCC lsa(A, C) F: GGCAATCGCTTGTGTTTTAGCG 55˚C [4] R: GTGAATCCCATGATGTTGATACC erm(A, B, F: GAAATIGGIIIIGGIAAAGGICA 37˚C [2] C, G) R: AATTGATTCTTIGTAAA msr(A, B) F: GCAAATGGTGTAGGTAAGACAACT 55˚C [2] R: ATCATGTGATGTAAACAAAAT 27F AGAGTTTGATCCTGGCTCAG 55˚C 1492R GGTTACCTTGTTACGACTT 55˚C 337F GACTCCTACGGGAGGCWGCAG 785F GGATTAGATACCCTGGTA M13 F: GTTTTCCCAGTCACGAC R: CAGGAAACAGCTATGAC 2
TABLE S2. Previously unpublished PCR primers used in this study Anneal GenBank accessions Target Primer sequence (5'-3') temp used for primer design ere(A, B) F: CTCATTTYRTMRMRGARTT 45˚C AY183453, A15097 R: GGWGTTTTTTGWAKATG erm(G, T) F: AAATATAAAAGATAGTCAAAA 45˚C L42817.1, M64090.1 R: CCATATTCCACTATTAAATAAG mph(A, B, C) F: TGGGTKCTRMGMWTSCCK 50˚C D16251, D85892, AB013298 R: ARCCCYTCTTCMCCAAA mph(D) F: CTCCTGTAACCAAGCCAATTG 55˚C AB048591 R: TTATCAACCCCGACCAGATTA mph(E) F: ATGACAATTCAAGATATTCAATC 50˚C FR751518 R: TTATATAACTCCCAACTGAGC mph(F) F: ATGCTGCACGACACGGACCG 55˚C AM260957 R: TCAAATCCCTGGCGCCGAC vat(A, C, F) F: ATTGGDGATAARYTRAT 45˚C L07778, AF015628, AF170730 R: ACMGGCATAATBRWYACATC vat(B, D, E) F: TTATYATGAAYGGWGCMAAYCA 50˚C U19459, L12033, AF139725 R: ATKGCWCCRTCHCCKATTT vat(H) F: ATGGCAGAAAAATTAAAAGG 45˚C GQ205627.2 R: CTAATCATTTTCTTTAGAAA vgb(B) F: GTTTCTATGCTGATCTGAATC 50˚C AF015628 R: GGTCTAAATGGCGATATATATGG 3
Table S3. GenBank accession numbers for amplified antibiotic resistance genesa GenBank Isolate and gene amplified accession KF245571 Enterococcus faecium F6S1 MsrC (msrC) gene KF245572 Pediococcus pentosaceus F4S4 MsrA (msrA) gene KF245573 Pediococcus pentosaceus F8S2 MsrA (msrA) gene KF245574 Enterococcus faecalis F5S8 Lsa (lsa) gene KF245575 Enterococcus faecalis F3S3 Lsa (lsa) gen KF245576 Streptococcus lutetiensis F5S7 rRNA methylase (ermTR) gene KF245577 Lactobacillus casei F4S1 rRNA methylase (ermG) gene KF245578 Pediococcus pentosaceus F7S1 rRNA methylase (ermB) gene KF245579 Pediococcus pentosaceus F4S2 rRNA methylase (ermB) gen KF245580 Enterococcus casseliflavus F4S7 rRNA methylase (ermB) gene KF245581 Lactobacillus fermentum F4S8 rRNA methylase (ermB) gene KF245582 Lactococcus lactis F4S9 rRNA methylase (ermB) gene KF245583 Pediococcus pentosaceus F4S10 rRNA methylase (ermB) gene KF245584 Lactobacillus fermentum F5S2 rRNA methylase (ermB) gene KF245585 Pediococcus pentosaceus F5S3 rRNA methylase (ermB) gene KF245586 Lactobacillus brevis F5S4 rRNA methylase (ermB) gene KF245587 Lactobacillus plantarum F5S5 rRNA methylase (ermB) gene KF245588 Enterococcus faecium F6S1 rRNA methylase (ermB) gene KF245589 Weissella confusa F2S1 rRNA methylase (ermB) gene KF245590 Weissella confusa F2S2 rRNA methylase (ermB) gene KF245591 Pediococcus pentosaceus F7S2 rRNA methylase (ermB) gene KF245592 Pediococcus pentosaceus F7S3 rRNA methylase (ermB) gene KF245593 Pediococcus pentosaceus F4S4 rRNA methylase (ermB) gene KF245594 Lactobacillus paracasei F4S3 rRNA methylase (ermG) gene aAmplified genes are partial sequence only 4
TABLE S4. DNA sequences amplified with vat PCR primersa Isolate vat gene sequence TTATTATGAACGGTGCCAATCACGTAATGAAAGGTATTTCAACTTATC CATTTAATATTTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTGA Pediococcus CTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTGGT pentosaceus F7S1 TTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATCGGGGACGGA GCCAT TTATTATGAACGGAGCAAATCACGTAATGAAAGGTATTTCAACTTATC CATTTAATATCTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTGA Pediococcus CTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTGGT pentosaceus F7S4 TTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATCGGAGACGGT GCCAT TTATTATGAACGGTGCCAACCACGTAATGAAAGGTATTTCGACTTATC CATTTAATATTTTAGGTGGCGATTGGCAACAATACACTCCTGAACTGA Pediococcus CTGATTTGCCGTTGAAAGGTGATACCGTAGTCGGAAGTGACGTGTGG pentosaceus F4S2 TTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATAGGGGACGG AGCCAT TTTATTATGAATGGTGCCAATCACGTAATGAAAGGTATTTCAACTTAT CCATTTAATATTTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTG Pediococcus ACTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTG pentosaceus F7S2 GTTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATAGGAGACG G TTTATTATGAACGGAGCCAATCACGTAATGAAAGGTATTTCAACTTAT CCATTTAATATTTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTG Pediococcus ACTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTG pentosaceus F7S3 GTTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATAGGAGACG G AAGGTATTTCGACTTATCCATTTAATATTTTAGGTGGCGATTGGCAAC Pediococcus AATACACTCCTGAACTGACTGATTTGCCGTTGAAAGGTGATACTGTAG pentosaceus F4S10 TCGGAAATGACGTGTGGTTTGGGCAAAATGTGACCGTCCTACCAGGC GTAAAAATAGGAGACGGAGCCAT TTTATTATGAACGGAGCCAACCACGTAATGAAAGGTATTTCAACTTAT CCATTTAATATTTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTG Lactobacillus plantarum ACTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTG F5S5 GTTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATAGGTGACG GTGC TTTATTATGAATGGAGCCAACCACGTAATGAAAGGTATTTCGACTTAT CCATTTAATATTTTAGGTGGCGATTGGCAACAATACACTCCTGAACTG Lactobacillus fermentum ACTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTG F4S8 GTTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATAGGAGACG GAGCCAT TTTATCATGAACGGAGCAAACCACGTAATGAAAGGTATTTCAACTTAT CCATTTAATATTTTGGGTGGCGATTGGCAGAAATATACTCCTGAACTG Pediococcus ACTGATTTGCCGTTGAAAGGTGATACTGTAGTCGGAAATGACGTGTG pentosaceus F1S2 GTTTGGGCAAAATGTGACCGTCCTACCAGGCGTAAAAATCGGAGACG GTGCCAT Isolate acetyltransferase gene sequence GGCATAATTGATACATCTTGACCTATCCAGACATCATTACCAATCACA GTATCTCCCTTAAAAGGAAGTTGTTCTATTGTAGGAGTCACTTTTCCC Lactococcus lactis F4S9 CATCCACAGCCAAAGATATTAAAGGGGTAGGTTGTAATTCCATCCATT CTATGATTTGCACCATTCATAATAAACTTAACACCCTCTGCAATTGCA CAGAACTTACCTATTATCAGTTTATCCCC aBecause amplified DNA sequences are <200 nt, they have not been submitted to GenBank 5
REFERENCES
1. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L (1996) Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40: 2562-2566. 2. Jalava J, Marttila H (2004) Application of molecular genetic methods in macrolide, lincosamide and streptogramin resistance diagnostics and in detection of drug-resistant Mycobacterium tuberculosis. APMIS 112: 838-855. 3. Lina G, Quaglia A, Reverdy M-E, Leclercq R, Vandenesch F, et al. (1999) Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother 43: 1062-1066. 4. Singh KV, Murray BE (2005) Differences in the Enterococcus faecalis lsa locus that influence susceptibility to quinupristin-dalfopristin and clindamycin. AntimicrobAgents Chemother 49: 32- 39.