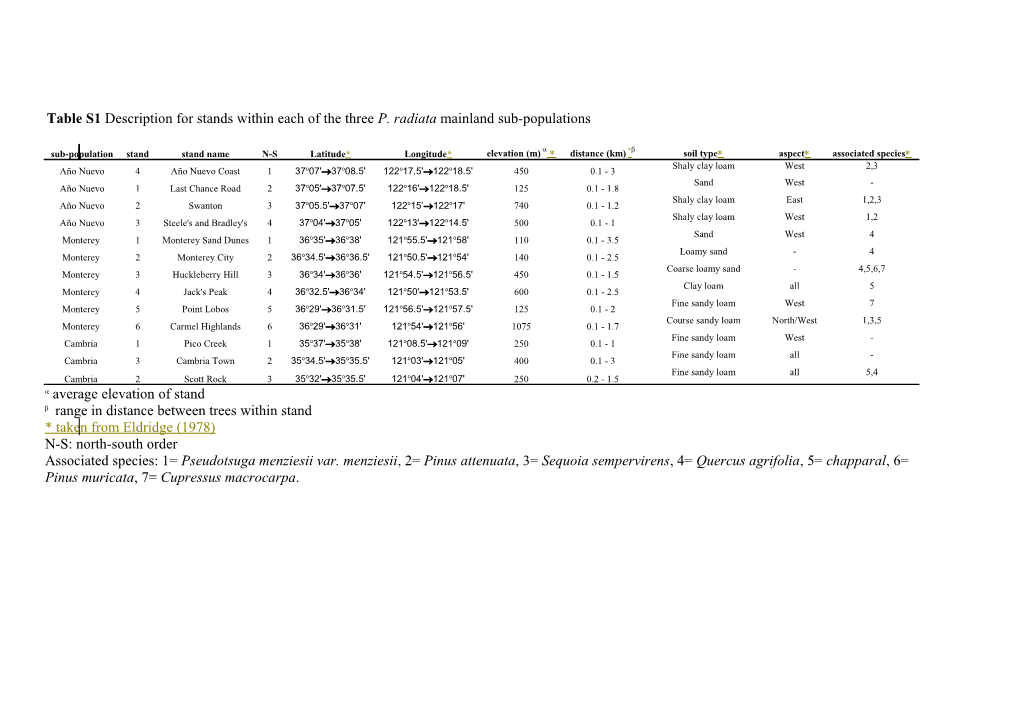
Table S1 Description for stands within each of the three P. radiata mainland sub-populations
sub-population stand stand name N-S Latitude* Longitude* elevation (m) distance (km) soil type aspect associated species Shaly clay loam West 2,3 Año Nuevo 4 Año Nuevo Coast 1 37o07'37o08.5' 122o17.5'122o18.5' 450 0.1 - 3 Sand West - Año Nuevo 1 Last Chance Road 2 37o05'37o07.5' 122o16'122o18.5' 125 0.1 - 1.8 Shaly clay loam East 1,2,3 Año Nuevo 2 Swanton 3 37o05.5'37o07' 122o15’122o17' 740 0.1 - 1.2 Shaly clay loam West 1,2 Año Nuevo 3 Steele's and Bradley's 4 37o04'37o05' 122o13'122o14.5' 500 0.1 - 1 Sand West 4 Monterey 1 Monterey Sand Dunes 1 36o35'36o38' 121o55.5'121o58' 110 0.1 - 3.5 Loamy sand - 4 Monterey 2 Monterey City 2 36o34.5'36o36.5' 121o50.5'121o54' 140 0.1 - 2.5 Coarse loamy sand - 4,5,6,7 Monterey 3 Huckleberry Hill 3 36o34'36o36' 121o54.5'121o56.5' 450 0.1 - 1.5 Clay loam all 5 Monterey 4 Jack's Peak 4 36o32.5'36o34' 121o50'121o53.5' 600 0.1 - 2.5 Fine sandy loam West 7 Monterey 5 Point Lobos 5 36o29'36o31.5' 121o56.5'121o57.5' 125 0.1 - 2 Course sandy loam North/West 1,3,5 Monterey 6 Carmel Highlands 6 36o29'36o31' 121o54'121o56' 1075 0.1 - 1.7 Fine sandy loam West - Cambria 1 Pico Creek 1 35o37'35o38' 121o08.5'121o09' 250 0.1 - 1 Fine sandy loam all - Cambria 3 Cambria Town 2 35o34.5'35o35.5' 121o03'121o05' 400 0.1 - 3 Fine sandy loam all 5,4 Cambria 2 Scott Rock 3 35o32'35o35.5' 121o04'121o07' 250 0.2 - 1.5 average elevation of stand range in distance between trees within stand * taken from Eldridge (1978) N-S: north-south order Associated species: 1= Pseudotsuga menziesii var. menziesii, 2= Pinus attenuata, 3= Sequoia sempervirens, 4= Quercus agrifolia, 5= chapparal, 6= Pinus muricata, 7= Cupressus macrocarpa. Table S2 Species richness and Sørensen’s similarity coefficient estimated for the three mainland populations
species present sub-population stand 0 1 2 3 4 5 6 7 SR pop. SR common sp. QS pop. QS Arbitrary population 1 1 1 1 1 5 Año Nuevo 1 1 1 1 0.33 2 1 1 1 1 4 3 0.67 3 1 1 1 3 2 0.50 4 1 1 1 3 2.75 2 0.50 0.500 Monterey 1 1 1 2 1 0.29 2 1 1 2 1 0.29 3 1 1 1 1 1 5 3 0.60 4 1 1 2 2 0.57 5 1 1 2 1 0.29 6 1 1 1 1 4 2.83 4 0.89 0.486 Cambria 1 1 1 1 0.33 2 1 1 1 0.33 3 1 1 1 3 1.67 2 0.50 0.389 1= Pseudotsuga menziesii var. menziesii, 2= Pinus attenuata, 3= Sequoia sempervirens, 4= Quercus agrifolia, 5= chapparal, 6= Pinus muricata, 7= Cupressus macrocarpa SR = species richness, total number of species recorded for each sub-population common sp. = number of species in common with an arbitrarily defined species (first row) for each sub population QS = Sørensen’s similarity coefficient: QS=2C/(A+B), where A and B are the number of species in samples A and B, respectively, and C is the number of species shared Arbitrary population = population with artificial species composition used as sample A when estimating Sørensen’s similarity coefficient by comparison with each actual population. pop. = population average for SR or QS Table S3 List of amplicons sequenced from 38 genes, from which 149 SNPs were selected for genotyping. gene name amplicon L SNPs F gene functional studies alpha expansin exp1-1F 307 2 4 (Darley et al. 2001) alpha expansin exp1-1R 307 3 4 (Darley et al. 2001) alpha expansin exp1-2 264 1 4 (Darley et al. 2001) alpha expansin exp1-4 614 1 4 (Darley et al. 2001) 2,4 (Wasteneys and Galway 2003; Wightman and alpha tubulin tub-1 788 2 Turner 2008) aquaporin porin-1 1919 5 4,5 (Kaldenhoff and Fischer 2006) At. COBL4 (cobra-like protein) cobl4-1 1200 1 2 (Roudier et al. 2005) At. COBL4 (cobra-like protein) cobl4-2 1442 3 2 (Roudier et al. 2005) At. dehydrin 2 dh2 606 2 5 (Caruso et al. 2002) At. dehydrin 7/8 dh7/8 232 2 5 (Caruso et al. 2002) At. rac13 GTPase rac13-1F 950 1 2 (Delmer et al. 1995; Potikha et al. 1999) At. rac13 GTPase rac13-2F 731 1 2 (Delmer et al. 1995; Potikha et al. 1999) At. rac13 GTPase rac13-3F 780 2 2 (Delmer et al. 1995; Potikha et al. 1999) At. rac13 GTPase rac13-4 911 2 2 (Delmer et al. 1995; Potikha et al. 1999) caffeate 3-O-methyltransferase2 comt2-1 645 1 1 (Allona et al. 1998) caffeate 3-O-methyltransferase2 comt2-2 645 1 1 (Allona et al. 1998) chloroplastic superoxide dismutase sodchl-1 720 3 5 (Edjolo et al. 2001; Rahman et al. 2004) cinnamate 4-hydroxylase c4h1-1 440 2 1 (Allona et al. 1998) cinnamoyl-CoA reductase ccr1-3 1611 1 1 (Peter and Neale 2004) cinnamoyl-CoA reductase promotor 1152 1 1 (Peter and Neale 2004) cinnamyl-alcohol dehydrogenase cad-1F 761 4 1 (Allona et al. 1998) cinnamyl-alcohol dehydrogenase cad-2F 750 5 1 (Allona et al. 1998) cinnamyl-alcohol dehydrogenase cad-3F 600 2 1 (Allona et al. 1998) endo-1,4- b-D-glucanase (korrigan) kor-2F 773 2 4 (Szyjanowicz et al. 2004) endo-1,4- b-D-glucanase (korrigan) kor-2R 850 2 4 (Szyjanowicz et al. 2004) endo-1,4- b-D-glucanase (korrigan) kor-3 900 1 4 (Szyjanowicz et al. 2004) Fragile fibre mutant 2 (katanin) fra2-2 786 3 2,4 (Burk et al. 2001) glycine hydroxymethyl transferase glyhmt-2F 650 3 1 (Vander Mijnsbrugge et al. 2000b) glycine hydroxymethyl transferase glyhmt-2R 650 2 1 (Vander Mijnsbrugge et al. 2000b) HDzip transcription factor (ATHB-8) athb-8-1 837 4 1,2 (Baima et al. 2001) HDzip transcription factor (ATHB-8) athb-8-2 784 3 1,2 (Baima et al. 2001) HDzip transcription factor (ATHB-X) athb-x-1 470 2 1,2 (Baima et al. 2001) peroxidase precursor perox-1 812 1 2 (Allona et al. 1998) phenylcoumaran benzylic ether reductase pcber-2 572 3 1 (Vander Mijnsbrugge et al. 2000a) proline rich protein prp1-1 643 6 3 (Zhang et al. 2000) Pt. cellulose synthase 1 cesa1-1F 893 3 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-1Fs 568 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-1R 292 2 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-2F 921 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-2Fs 827 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-2R 835 2 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-3F 860 2 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-3R 808 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-4 982 5 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 1 cesa1-5F 663 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 2 cesa2-4 716 1 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 3 cesa3-2F 800 2 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 3 cesa3-3R 708 3 2 (Nairn and Haselkorn 2005) Pt. cellulose synthase 3 cesa3-4F 736 1 2 (Nairn and Haselkorn 2005) Pt. lim 1 transcription factor lim1-1F 849 1 1 (Kawaoka et al. 2006; Kawaoka et al. 2001) Pt. lim 1 transcription factor lim1-2 853 1 1 (Kawaoka et al. 2006; Kawaoka et al. 2001) Pt. myb 1 transcription factor myb1-2 1361 2 1 (Kawaoka et al. 2006; Kawaoka et al. 2001) Pt. myb 4 transcription factor myb4-1F 717 1 1 (Patzlaff et al. 2003) Pt. myb 4 transcription factor myb4-2F 539 1 1 (Patzlaff et al. 2003) Pt. phenylalanine ammonia lyase 1 pal1-1 761 1 1 (Allona et al. 1998) Pt. phenylalanine ammonia lyase 1 pal1-3 503 4 1 (Allona et al. 1998) Pt. phenylalanine ammonia lyase 1 pal2-5 864 1 1 (Allona et al. 1998) Pt.arabinogalactan protein 4 agp4-1b 1049 1 3 (Zhang et al. 2000) Pt.arabinogalactan protein 4 agp4-2 700 5 3 (Zhang et al. 2000) Pt.arabinogalactan protein 6 agp6-1 497 1 3 (Zhang et al. 2000) putative cell-wall protein lp5-1 735 1 5 (Chang et al. 1996) S-adenosyl-L-homocysteine hydrolase sahh1-1 650 3 5 (Moffatt and Weretilnyk 2001) S-adenosylmethionine transferase sam1-3 605 2 1 (Moffatt and Weretilnyk 2001) S-adenosylmethionine transferase1 sam1-4 737 4 1 (Moffatt and Weretilnyk 2001) S-adenosylmethionine transferase2 sam2-1 685 1 1 (Moffatt and Weretilnyk 2001) S-adenosylmethionine transferase2 sam2-4 714 5 1 (Moffatt and Weretilnyk 2001) sucrose synthase susy-1F 593 1 2 (Doblin et al. 2002) sucrose synthase susy-1R 450 1 2 (Doblin et al. 2002) unknown protein nh3702 287 1 n/a n/a xyloglucan endotransglycosylase xet1-1 878 2 4 (Verbelen et al. 2001) xyloglucan endotransglycosylase xet1-2 887 1 4 (Verbelen et al. 2001) xet1- xyloglucan endotransglycosylase promotor 563 2 4 (Verbelen et al. 2001) TOTAL 54188 149 L= length of sequenced region F = functional category where: 1) lignin biosynthesis and deposition, 2) cellulose biosynthesis and deposition, 3) cell wall structure, 4) cell expansion, and 5) plant water relations/ abiotic stress responses
Table S4 Amplicons and their respective primer sequences Amplicon F primer (5'-3') R primer (5'-3') L exp1-2 TCTCGGTTCCCTTCCCTCAGTA GCTTCACGCACGGCACTCT 264 agp4-1b AAACTAGCCGAGCCATCAAGC TTTCTTCTGCTGCCGTTTCAA 1049 agp6-1 TCAGGGTCAACAATGGCGTTC GGGCTTTTCAGTGCGGACG 497 agp6-promotor TCACCACTCAACATCTGCAT GCCATTGTTGAACCTGAAAA 830 athb-8-1 GCACTTACTCTTGCCCGTTGG AGTGCTGCTTACAAAATCCATCCA 837 tub-1 GGTGGTATTGCAGGAGGCGT AGTTGTTGGCCGCATCTTCC 788 tub-promotor ACTATAGGGGCAGCGTGGT AAGACGCCTCCTGCAATAC 870 cad-1sdF GGATACTGAAAAGATGATGGTG CCCAAGAAGAAAGGACATACT 761 ccr1-3 AGTGGCCGAGAAGGCTGC GGAGTTATTTGGTGACAACATTATTGGTA 1611 ccr1-promotor GATTTTCTCATAATGAACTTAAGCA ATTAATCAAGAGGGCGATGATATTC 1152 cesa1-2R TTGCTTTAATTGTACTGGATTGG CACCGGCCTTCTTATGGT 835 cesa2-4 CAGTTCTGGGTCATTGGAG ATGAACACAGCCCCAATTT 716 cesa3-4F GCTCACCAATGCTCCATTTA CACAAGGTTGATGACAAGCA 736 cobl4-2 CAAGTTAGTGTGGGCAATTCAG GCAGATCATTATAATATTTCTGTCC 1442 fra1-1 CAACCGGAGTTGCTTTAGCC GAAAGTCTACAAGGGATTGTGAAGA 815 fra2-2 AGCACATCATCCCACCGTACT TCTTGGGTATGAAAGATTGC 786 kor-2F ATCTTGGCGAGGAAACTCTTGCAT AGACGACAATAGATCAGCCTGCAAC 773 lim1-1F AAAACCAGCAAAAACCAAACT TTCAGCTGCATAATCACTCG 849 lim1-1R AAAACCAGCAAAAACCAAACT TTCAGCTGCATAATCACTCG 745 myb4-1F GCCATTCCGTTCATTCAG AGATTTACTGTTGTTGGTGGAG 717 pal1-3 TCTTTACCTGGCAAATCCTG CGAGCTGGTTTCTCACAAAT 503 perox-1 CGACATAGCTGGAGGGCCAT GCTGCTTGCATGAGAAATTGC 812 porin-1 GAAATGGAAGGAAAAGAAGAAGA GAGCGAGTTTTGAAGGGAAT 1919 prp1-1 G1 TTCCTCCAATTCCTTGCACTG GGCGCTTGCTGTCAGAAAC 643 sam1-4 GCTCTCAGCCCATTGTTTAG GCAGGTTCCATTCTTTCTGA 737 sam2-4 TTCCCAGGATGTGTTTGTCT TAACCAGCACTTTGCAGTGA 714 susy-1F G1 CCTATTATCTTTTCTATGGCAAG TGTCTCGCGCCTTTCAAGT 593 xet1-1 CCATAAGCCCTCCCTTTCTA GAAGTCCAGAGCACGGAGTA 878 xet1-promotor CTCAATAGGTATGTTTTATATAGGG CCACCTGATTTCCATTGTGT 563 exp1-4 TCTCCCTCTCCTGCATTTTG AAACAACCCGCATTTCAAGA 614 exp1-F1 TCTCGGTTCCCTTCCCTCAGTA GCTTCACGCACGGCACTCT 307 exp1-R1 TCTCGGTTCCCTTCCCTCAGTA GCTTCACGCACGGCACTCT 307 agp4-2 TGAAGTTCCCGCCGAAAGC GATCCCCTGAAATCCGTTTACTACATAC 700 athb-8-2 AGGAAGCTTCTCGTCTCCAG AGTGCTGCTTACAAAATCCATCCA 784 athb-x-1 AAGTCATTGCCTGTTTTCACCTTTG GCTGCTTACATGCTCTCCCTCTG 470 c4h1-1 TGAACGCGGGAGGTTGG GCGAATCTGTAGTGCTGGACAA 440 cad-2F TTGTTTCATAGTTTGTGTTTTT TGATTTGATTGATTGATTGC 750 cad-3F GAATGATTGGAAAGTGACACAC CAGATTCATCCCAAAATGGAT 600 cesa1-1F CTTCCAACGGGACTATGAACT GGTACTCACGAAGAAGTCCAC 893 cesa1-1Fs CTTCCAACGGGACTATGAACT GGTACTCACGAAGAAGTCCAC 568 cesa1-1R CTTCCAACGGGACTATGAACT GGTACTCACGAAGAAGTCCAC 292 cesa1-2F TTAAGGTACGTACGAGAGACC CGTTTCTCACGAGAGACATAGA 921 cesa1-2Fs TTAAGGTACGTACGAGAGACC CGTTTCTCACGAGAGACATAGA 827 cesa1-3F ATGAATGCATTGGTTCGAGT GCTTCTTTGATGAGTGAAGCTG 860 cesa1-3R ATGAATGCATTGGTTCGAGT GCTTCTTTGATGAGTGAAGCTG 808 cesa1-4 AAAACCGAATGGGGGAAAG GACTCCTCCAATCACCCAAA 982 cesa1-5F TTGCAGTTTTTCAAGGTCTTC CCACCCGACCTGAAATAATC 663 cesa3-2F GGTGAGTTTGGGGAACTGTATG CACAATTTCAAACACAAATATGAGA 800 cesa3-3R CATGGATCCAAACAGGGATA GCGATCAATACCATCGAATC 708 cobl4-1 CTAATTTCCATTTTACATGTTTAAATTTTG AACTTGACAGTTTTGTTAGTGGTCCTT 1200 comt2-1 CGAAATGGGATCGGCTTCC CCTGAACCTCCTTAAAACCCTCG 645 Amplicon F primer (5'-3') R primer (5'-3') L comt2-2 TCCCCCACATTTCAGGAATCAAC AGGATCCACTGCAATCAAACA 645 dh2-1 GAAGAAGCACCAGAGCACAA ATTCATAACTTTAATCGACATAGGA 606 dh78-1 CCGCGGTATGTTCGGCTTAT ACAACCAGTCTTGTTTTGTTGTC 232 glyhmt2F1 AAGCAAGGTCTGTGGATCTCCG GGCTGTGTAGGCTGCGAAGTTAG 650 glyhmt2R1 AAGCAAGGTCTGTGGATCTCCG GGCTGTGTAGGCTGCGAAGTTAG 650 kor -2R ATCTTGGCGAGGAAACTCTTGCAT AGACGACAATAGATCAGCCTGCAAC 850 kor -3 GTCGTCTTTCCAGGAGGTTTG ATCTGCGTCGATGTAGCTTGA 900 lim1-2 ATCACCATGCTCAGCTCTTC GTACCAAATCCAAGGGCAAT 853 lp5-1 TGTGCCGCCGTTGAGAGCCGTA TATGGCCACACAATACACTGTAG 735 myb1-2 GCAACCGTGCTGTGACAA ATCAAGAAAGGGCCCCAGT 1361 myb4-2F CGGACAACGAGATCAAGAAT AAGATGCCGTGGAAGATACA 539 nh3702 TAAACCAAAGACGGCTGGAT CATTCTTTGCTTCGACTATTGTT 287 pal1-1 TGCCAGCTACCCATTGACTCAG TGATCTGCCACCCTTACATATTTCTG 761 pal2-5 ATTCCATTTGAACCCATCTG GGAGAACCTTTTCCTGCTTC 864 pcber-2 GGGTATTTCCTGCGAAGCCTC CGACGGTGGTGTATTTCACATCA 572 rac13-1F TTGGGTTCCTGCAGAGGTAT TGTCCCAGAGGCCAAGAT 950 rac13-2F TTAAGGTACGTACGAGAGACC CGTTTCTCACGAGAGACATAGA 731 rac13-3F ATGAATGCATTGGTTCGAGT GCTTCTTTGATGAGTGAAGCTG 780 rac13-4 ATTTGAAGAAGCAAATCG ATACAGACAAACTCGGCAAT 911 sahh1-1 TGGGTGCCAAGCTAACAAAGC TTGATTCATAACATGCCAGTGCTG 650 sam1-3 CGTTGACTCGTATGGTACAGGGAC GCTTAGGCATTACAGCCCACAGA 605 sam2-1 CATTCTCTGATTTGCCGTGCTG TCTTTCTGACCTCGGTGAGCTTC 685 sodchl1-6 GAAAATGTACATGTGTGGTAC GGTCAGAGAAAAAAATGTTAC 720 susy-1R CCTATTATCTTTTCTATGGCAAG TGTCTCGCGCCTTTCAAGT 450 tub-promotor ACTATAGGGGCAGCGTGGT AAGACGCCTCCTGCAATAC 870 xet1-2 TTGAATTTCTGGGAAACCTG TGTGTGCCTGAAAAAGAATG 887 xet1-promotor CTCAATAGGTATGTTTTATATAGGG CCACCTGATTTCCATTGTGT 563 Table S5 Significance of population pairwise comparisons (T-test) and phenotypic differentiation (PST) based on trait means and variances in the three mainland populations of radiata pine.
Trait Description Compartment A v. M A v. C M V. C A v. M A v. C M V. C 6 cumulative ring width whole core 0.394 0.000 0.000 -0.014 0.842 0.877 20 MFA whole core 0.448 0.057 0.069 -0.040 0.520 0.513 42 MOE corewood 0.239 0.029 0.085 0.151 0.575 0.444 24 ring width corewood 0.175 0.000 0.000 0.247 0.837 0.824 25 ring width outerwood 0.347 0.000 0.001 0.024 0.794 0.829 5 ring width whole core 0.498 0.002 0.001 -0.048 0.766 0.815 17 coarseness whole core 0.084 0.357 0.092 0.427 0.002 0.460 26 cumulative ring width corewood 0.038 0.281 0.022 0.558 0.075 0.631 8 density corewood 0.011 0.313 0.081 0.672 0.036 0.442 12 late wood density whole core 0.047 0.437 0.076 0.543 -0.057 0.518 19 specific surface area whole core 0.042 0.461 0.094 0.547 -0.065 0.471 30 tangential diameter corewood 0.047 0.389 0.041 0.530 -0.034 0.533 18 wall thickness whole core 0.023 0.446 0.093 0.615 0.060 0.460 7 density whole core 0.023 0.137 0.317 0.618 0.035 0.349 2 late wood density outerwood 0.005 0.150 0.216 0.723 0.296 0.240 38 specific surface area corewood 0.035 0.333 0.139 0.573 0.012 0.328 36 wall thickness corewood 0.016 0.224 0.148 0.645 0.147 0.308 37 wall thickness outerwood 0.016 0.112 0.339 0.641 0.375 0.051 9 density outerwood 0.036 0.067 0.425 0.504 0.545 -0.009 23 early wood density outerwood 0.008 0.005 0.175 0.689 0.732 0.297 11 early wood density whole core 0.014 0.086 0.358 0.657 0.422 0.033 39 specific surface area outerwood 0.021 0.045 0.417 0.617 0.552 -0.005 31 tangential diameter outerwood 0.097 0.020 0.137 0.404 0.623 0.346 15 tangential diameter whole core 0.025 0.038 0.398 0.603 0.557 0.004 27 cumulative ring width outerwood 0.057 0.000 0.000 -0.046 0.827 0.867 41 MFA outerwood 0.020 0.000 0.000 0.626 0.880 0.871 43 MOE outerwood 0.003 0.000 0.001 0.742 0.866 0.805 21 MOE whole core 0.053 0.002 0.043 0.510 0.754 0.592 22 early wood density corewood 0.056 0.459 0.107 0.498 -0.065 0.380 35 coarseness outerwood 0.035 0.250 0.238 0.423 0.234 0.314 1 late wood density corewood 0.315 0.332 0.198 0.034 0.059 0.212 14 radial diameter whole core 0.167 0.304 0.424 0.243 0.100 -0.029 16 cell population whole core 0.272 0.222 0.370 0.082 0.218 0.007 28 radial diameter corewood 0.216 0.092 0.216 0.234 0.115 0.-0.03 29 radial diameter outerwood 0.117 0.238 0.448 0.338 0.196 -0.039 32 cell population corewood 0.285 0.248 0.134 0.065 0.176 0.343 33 cell population outerwood 0.437 0.223 0.255 -0.058 0.218 0.140 34 coarseness corewood 0.067 0.186 0.374 0.415 0.205 0.386 40 MFA corewood 0.440 0.311 0.265 -0.059 0.083 0.114 p-value for pairwise t-test of phenotype difference between population pair FST-PST for population pairs MFA = cellulose microfibril angle MOE = modulus of elasticity (stiffness) Table S6 Principal component loadings for climate variables
Climate CLIMPCA rainfall 1.000 evaporation -0.996 temp -0.830 % of Var. 99
% of Var. = total percentage of variation in the data set explained by the principal component.
Table S7 Principal component loadings for ecological variables
Ecology ECOLPCA Species richness 0.993 Species similarity 0.993 % of Var. 98.5
% of Var. = total percentage of variation in the data set explained by the principal component.
Table S8 Principal component loadings for individual outlier SNPs
SNP SNPPCA1 SNPPCA2 9 0.557 0.222 16 -0.616 -0.082 57 -0.617 -0.171 59 0.664 0.016 80 0.509 0.295 108 0.685 0.050 139 0.556 -0.071 2 -0.323 0.685 70 0.299 -0.714 % of Var. 30.479 12.886
% of Var. = total percentage of variation in the data set explained by the principal component. For each SNP the major component loading is highlighted in bold. Table S9 Principal component loadings for individual traits
Trait Trait TRAITPCA1 TRAITPCA2 trait type 5 ring width -0.462 0.76 growth 27 cumulative ring width -0.407 0.796 growth 25 ring width -0.391 0.654 growth 6 cumulative ring width -0.39 0.789 growth 24 ring width -0.37 0.622 growth 31 tangential diameter -0.257 0.626 growth 15 tangential diameter -0.248 0.597 growth 26 cumulative ring width -0.193 0.539 growth 30 tangential diameter -0.163 0.427 growth 39 specific surface area -0.795 -0.405 wood properties 19 specific surface area -0.777 -0.558 wood properties 38 specific surface area -0.564 -0.528 wood properties 41 MFA -0.537 0.277 wood properties 20 MFA -0.466 0.058 wood properties 17 coarseness 0.377 0.799 wood properties 42 MOE 0.536 0.088 wood properties 36 wall thickness 0.608 0.507 wood properties 8 density 0.676 0.22 wood properties 2 late wood density 0.733 -0.001 wood properties 12 late wood density 0.781 0.103 wood properties 21 MOE 0.812 -0.103 wood properties 23 early wood density 0.814 -0.218 wood properties 18 wall thickness 0.815 0.504 wood properties 37 wall thickness 0.816 0.39 wood properties 43 MOE 0.818 -0.241 wood properties 11 early wood density 0.842 -0.096 wood properties 9 density 0.907 -0.067 wood properties 7 density 0.929 0.062 wood properties % of Var. 39.97 22.17
The traits in this table were defined as putatively adaptive following estimation of PST. % of Var. = total percentage of variation in the data set explained by the principal component. Trait type = traits grouped broadly as either wood or growth traits, a detailed description of each trait is presented in table S5. For each trait the major component loading is highlighted in bold. Table S10 Matrices of P-values for ANOVA comparing principal component variation for outlier SNPs and traits among the three mainland populations (environmental regimes)
Año Nuevo Monterey Cambria
A TRAITPCA1 1
M TRAITPCA1 0.015 1
C TRAITPCA1 0.003 0.289 1
A TRAITPCA2 1
M TRAITPCA2 0.680 1
C TRAITPCA2 0.004 0.001 1
A SNPPCA1 1
M SNPPCA1 4.57E-78 1
C SNPPCA1 6.95E-58 2.14E-07 1
A SNPPCA2 1
M SNPPCA2 1.2E-16 1
C SNPPCA2 6.35E-18 2.83E-41 1 A: Año Nuevo M: Monterey C: Cambria st TRAITPCA1: 1 principle component for trait data nd TRIATPCA2: 2 principle component for trait data st SNPPCA1: 1 principle component for outlier SNP data nd SNPPCA2: 2 principle component for outlier SNP data Table S11 Pair-wise estimates of genetic differentiation (FST), between stands within populations using putatively neutral non-outlier SNPs.
Año Nuevo Stand 1 Stand2 Stand3 Stand4 Stand1 0 Stand2 0.000* 0 Stand3 0.014** 0.018* 0 Stand4 0.021** 0.016* 0.025** 0
Monterey Stand 1 Stand2 Stand3 Stand4 Stand5 Stand6 Stand1 0 Stand2 0.004 0 Stand3 0.000 0.001 0 Stand4 0.002 0.003 0.002 0 Stand5 0.000 0.008 0.000 0.000 0 Stand6 0.000 0.002 0.000 0.000 0.000 0
Cambria Stand 1 Stand2 Stand3 Stand1 0 Stand2 0.003 0 Stand3 0.000 0.001 0
* significant at P ≤ 0.05, based on 999 permutations ** significant at P ≤ 0.01, based on 999 permutations *** significant at P ≤ 0.001, based on 999 permutations