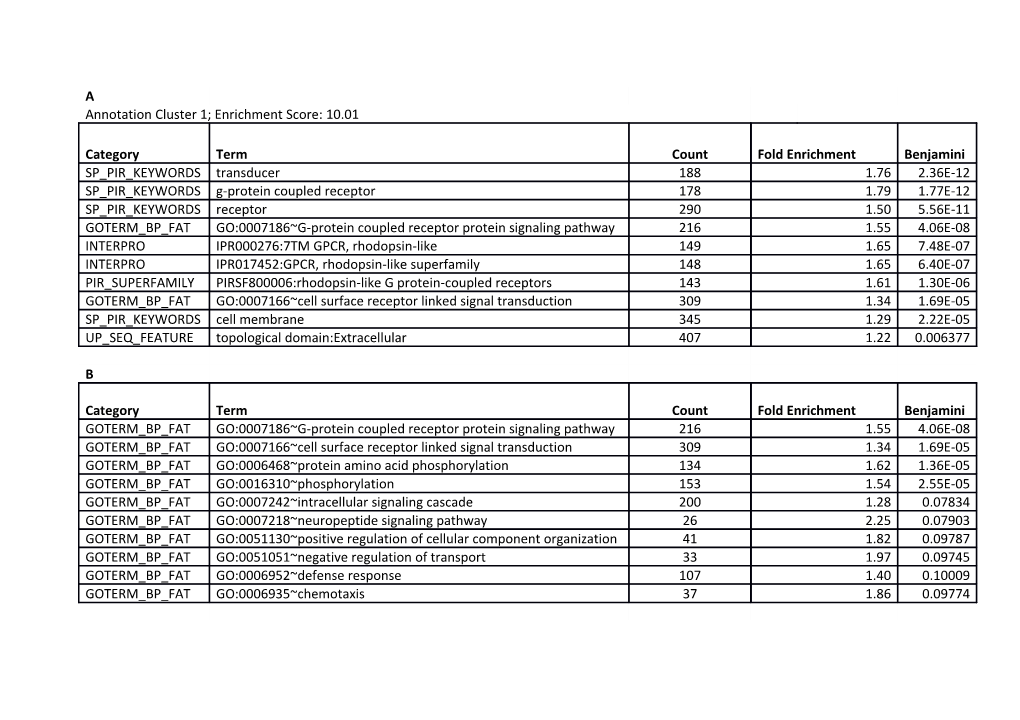
A Annotation Cluster 1; Enrichment Score: 10.01
Category Term Count Fold Enrichment Benjamini SP_PIR_KEYWORDS transducer 188 1.76 2.36E-12 SP_PIR_KEYWORDS g-protein coupled receptor 178 1.79 1.77E-12 SP_PIR_KEYWORDS receptor 290 1.50 5.56E-11 GOTERM_BP_FAT GO:0007186~G-protein coupled receptor protein signaling pathway 216 1.55 4.06E-08 INTERPRO IPR000276:7TM GPCR, rhodopsin-like 149 1.65 7.48E-07 INTERPRO IPR017452:GPCR, rhodopsin-like superfamily 148 1.65 6.40E-07 PIR_SUPERFAMILY PIRSF800006:rhodopsin-like G protein-coupled receptors 143 1.61 1.30E-06 GOTERM_BP_FAT GO:0007166~cell surface receptor linked signal transduction 309 1.34 1.69E-05 SP_PIR_KEYWORDS cell membrane 345 1.29 2.22E-05 UP_SEQ_FEATURE topological domain:Extracellular 407 1.22 0.006377
B
Category Term Count Fold Enrichment Benjamini GOTERM_BP_FAT GO:0007186~G-protein coupled receptor protein signaling pathway 216 1.55 4.06E-08 GOTERM_BP_FAT GO:0007166~cell surface receptor linked signal transduction 309 1.34 1.69E-05 GOTERM_BP_FAT GO:0006468~protein amino acid phosphorylation 134 1.62 1.36E-05 GOTERM_BP_FAT GO:0016310~phosphorylation 153 1.54 2.55E-05 GOTERM_BP_FAT GO:0007242~intracellular signaling cascade 200 1.28 0.07834 GOTERM_BP_FAT GO:0007218~neuropeptide signaling pathway 26 2.25 0.07903 GOTERM_BP_FAT GO:0051130~positive regulation of cellular component organization 41 1.82 0.09787 GOTERM_BP_FAT GO:0051051~negative regulation of transport 33 1.97 0.09745 GOTERM_BP_FAT GO:0006952~defense response 107 1.40 0.10009 GOTERM_BP_FAT GO:0006935~chemotaxis 37 1.86 0.09774 C
Category Term Count Fold Enrichment Benjamini GOTERM_MF_FAT GO:0004672~protein kinase activity 128 1.73 3.51E-07 GOTERM_MF_FAT GO:0001653~peptide receptor activity 40 2.88 4.76E-07 GOTERM_MF_FAT GO:0008528~peptide receptor activity, G-protein coupled 40 2.88 4.76E-07 GOTERM_MF_FAT GO:0004674~protein serine/threonine kinase activity 94 1.79 6.56E-06 GOTERM_MF_FAT GO:0008188~neuropeptide receptor activity 19 3.90 8.70E-05 GOTERM_MF_FAT GO:0042923~neuropeptide binding 19 3.80 1.10E-04 GOTERM_MF_FAT GO:0042277~peptide binding 50 2.02 3.55E-04 GOTERM_MF_FAT GO:0001608~nucleotide receptor activity, G-protein coupled 14 3.96 0.00248 GO:0045028~purinergic nucleotide receptor activity, G-protein GOTERM_MF_FAT coupled 14 3.96 0.00248 GOTERM_MF_FAT GO:0001614~purinergic nucleotide receptor activity 14 3.19 0.03203 GOTERM_MF_FAT GO:0016502~nucleotide receptor activity 14 3.19 0.03203
D Annotation Cluster 1; Enrichment score: 12.14
Category Term Count Fold Enrichment Benjamini SP_PIR_KEYWORDS g-protein coupled receptor 119 2.56 1.03E-18 SP_PIR_KEYWORDS transducer 124 2.49 6.94E-19 GOTERM_BP_FAT GO:0007186~G-protein coupled receptor protein signaling pathway 141 2.17 9.01E-16 SP_PIR_KEYWORDS receptor 172 1.91 7.06E-15 INTERPRO IPR000276:7TM GPCR, rhodopsin-like 98 2.31 1.33E-11 INTERPRO IPR017452:GPCR, rhodopsin-like superfamily 97 2.29 1.63E-11 GOTERM_BP_FAT GO:0007166~cell surface receptor linked signal transduction 181 1.68 4.75E-10 PIR_SUPERFAMILY PIRSF800006:rhodopsin-like G protein-coupled receptors 94 2.06 2.51E-09 UP_SEQ_FEATURE topological domain:Cytoplasmic 274 1.42 8.61E-07 KEGG_PATHWAY hsa04080:Neuroactive ligand-receptor interaction 45 2.54 1.29E-06 SP_PIR_KEYWORDS cell membrane 187 1.50 1.31E-06 UP_SEQ_FEATURE topological domain:Extracellular 222 1.42 2.71E-05 GOTERM_CC_FAT GO:0005886~plasma membrane 290 1.26 5.96E-04 GOTERM_CC_FAT GO:0016021~integral to membrane 363 1.13 0.12542
E Annotation Cluster 1; Enrichment Score: 2.61
Category Term Count Fold Enrichment Benjamini GOTERM_MF_FAT GO:0004672~protein kinase activity 65 1.68 0.04 SP_PIR_KEYWORDS kinase 73 1.62 0.03 GOTERM_BP_FAT GO:0006468~protein amino acid phosphorylation 71 1.60 0.24 SP_PIR_KEYWORDS serine/threonine-protein kinase 46 1.85 0.02 UP_SEQ_FEATURE active site:Proton acceptor 69 1.60 0.33 UP_SEQ_FEATURE domain:Protein kinase 53 1.72 0.20 GOTERM_MF_FAT GO:0004674~protein serine/threonine kinase activity 48 1.75 0.09 INTERPRO IPR017441:Protein kinase, ATP binding site 50 1.67 0.47 GOTERM_BP_FAT GO:0016310~phosphorylation 78 1.47 0.60 INTERPRO IPR008271:Serine/threonine protein kinase, active site 41 1.76 0.34 UP_SEQ_FEATURE nucleotide phosphate-binding region:ATP 90 1.42 0.37 INTERPRO IPR000719:Protein kinase, core 51 1.63 0.28 INTERPRO IPR017442:Serine/threonine protein kinase-related 41 1.74 0.23 INTERPRO IPR002290:Serine/threonine protein kinase 30 1.76 0.63 UP_SEQ_FEATURE binding site:ATP 53 1.49 0.86 SMART SM00220:S_TKc 30 1.71 0.75 SP_PIR_KEYWORDS transferase 115 1.26 0.48 SP_PIR_KEYWORDS nucleotide-binding 135 1.23 0.49 SP_PIR_KEYWORDS atp-binding 109 1.26 0.45 GOTERM_MF_FAT GO:0000166~nucleotide binding 169 1.18 0.93 GOTERM_MF_FAT GO:0005524~ATP binding 115 1.22 0.95 GOTERM_MF_FAT GO:0032559~adenyl ribonucleotide binding 116 1.21 0.91 GOTERM_MF_FAT GO:0032555~purine ribonucleotide binding 139 1.19 0.90 GOTERM_MF_FAT GO:0032553~ribonucleotide binding 139 1.19 0.90 GOTERM_MF_FAT GO:0017076~purine nucleotide binding 142 1.16 0.97 GOTERM_BP_FAT GO:0006796~phosphate metabolic process 80 1.24 0.92 GOTERM_BP_FAT GO:0006793~phosphorus metabolic process 80 1.24 0.92 GOTERM_MF_FAT GO:0030554~adenyl nucleotide binding 118 1.17 0.96 GOTERM_MF_FAT GO:0001883~purine nucleoside binding 119 1.16 0.95 GOTERM_MF_FAT GO:0001882~nucleoside binding 119 1.16 0.96
Annotation Cluster 2; Enrichment Score: 2.18
Category Term Count Fold Enrichment Benjamini GOTERM_BP_FAT GO:0051674~localization of cell 34 1.66 0.86 GOTERM_BP_FAT GO:0048870~cell motility 34 1.66 0.86 GOTERM_BP_FAT GO:0006928~cell motion 47 1.49 0.90 GOTERM_BP_FAT GO:0016477~cell migration 29 1.58 0.91
Annotation Cluster 3; Enrichment Score: 1.97 Category Term Count Fold Enrichment Benjamini GOTERM_BP_FAT GO:0007163~establishment or maintenance of cell polarity 10 3.07 0.84 GO:0035088~establishment or maintenance of apical/basal cell GOTERM_BP_FAT polarity 5 5.01 0.91 GO:0045197~establishment or maintenance of epithelial cell GOTERM_BP_FAT apical/basal polarity 4 6.68 0.91
Annotation Cluster 4; Enrichment Score: 1.86
Category Term Count Fold Enrichment Benjamini INTERPRO IPR017907:Zinc finger, RING-type, conserved site 31 1.68 0.69 INTERPRO IPR001841:Zinc finger, RING-type 32 1.64 0.71 SMART SM00184:RING 32 1.60 0.60 UP_SEQ_FEATURE zinc finger region:RING-type 24 1.74 0.99 INTERPRO IPR018957:Zinc finger, C3HC4 RING-type 21 1.40 1.00
Annotation Cluster 5; Enrichment Score: 1.84
Category Term Count Fold Enrichment Benjamini GOTERM_CC_FAT GO:0016323~basolateral plasma membrane 25 1.89 0.80 GOTERM_CC_FAT GO:0030055~cell-substrate junction 16 2.19 0.76 GOTERM_CC_FAT GO:0005925~focal adhesion 15 2.26 0.64 GOTERM_CC_FAT GO:0005924~cell-substrate adherens junction 15 2.17 0.57 GOTERM_CC_FAT GO:0005912~adherens junction 19 1.88 0.62 GOTERM_CC_FAT GO:0070161~anchoring junction 19 1.70 0.75 GOTERM_CC_FAT GO:0030054~cell junction 37 1.10 0.97
Supplementary Table S2. DAVID gene ontology analysis. (A) Terms in the top functional cluster in list SL1. (B) Enrichment for biological processes in list SL1 (top ten terms). (C) Significant molecular function terms in list SL1 after Benajmini correction. (D) Terms in the top functional cluster in list SL2. (E) Top five functional clusters in candidates with 50-85% reduction in MCF10A viability.