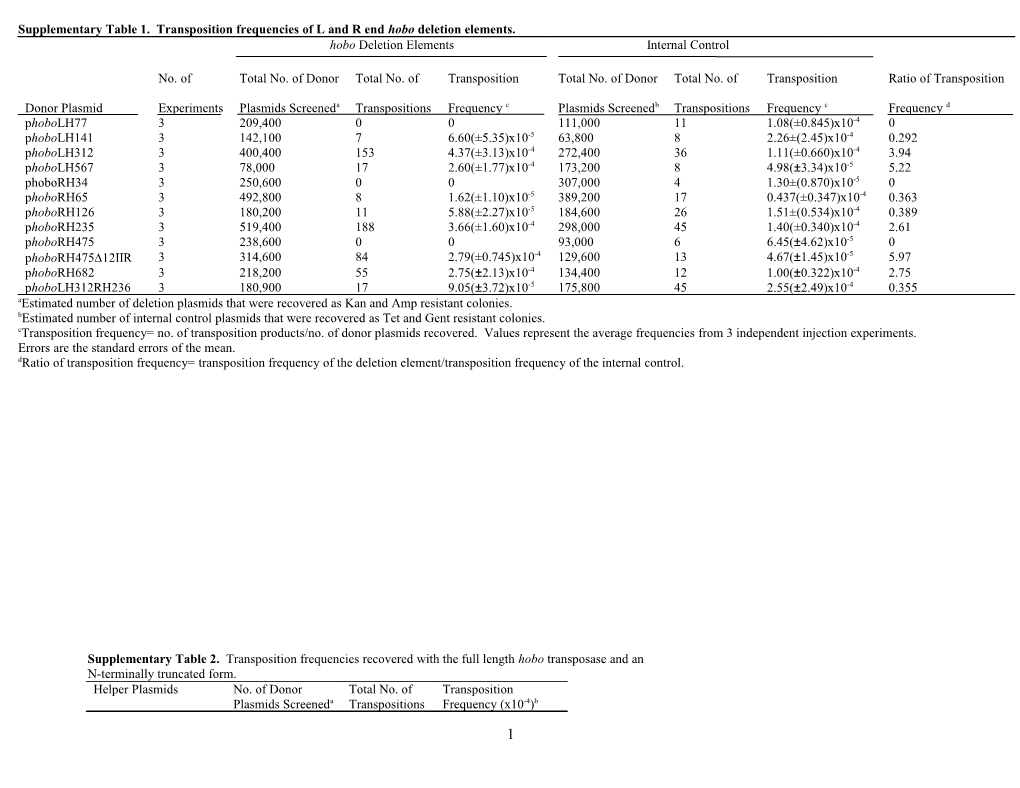
Supplementary Table 1. Transposition frequencies of L and R end hobo deletion elements. hobo Deletion Elements Internal Control
No. of Total No. of Donor Total No. of Transposition Total No. of Donor Total No. of Transposition Ratio of Transposition
Donor Plasmid Experiments Plasmids Screeneda Transpositions Frequency c Plasmids Screenedb Transpositions Frequency c Frequency d phoboLH77 3 209,400 0 0 111,000 11 1.08(±0.845)x10-4 0 phoboLH141 3 142,100 7 6.60(±5.35)x10-5 63,800 8 2.26±(2.45)x10-4 0.292 phoboLH312 3 400,400 153 4.37(±3.13)x10-4 272,400 36 1.11(±0.660)x10-4 3.94 phoboLH567 3 78,000 17 2.60(±1.77)x10-4 173,200 8 4.98(±3.34)x10-5 5.22 phoboRH34 3 250,600 0 0 307,000 4 1.30±(0.870)x10-5 0 phoboRH65 3 492,800 8 1.62(±1.10)x10-5 389,200 17 0.437(±0.347)x10-4 0.363 phoboRH126 3 180,200 11 5.88(±2.27)x10-5 184,600 26 1.51±(0.534)x10-4 0.389 phoboRH235 3 519,400 188 3.66(±1.60)x10-4 298,000 45 1.40(±0.340)x10-4 2.61 phoboRH475 3 238,600 0 0 93,000 6 6.45(±4.62)x10-5 0 phoboRH47512IIR 3 314,600 84 2.79(±0.745)x10-4 129,600 13 4.67(±1.45)x10-5 5.97 phoboRH682 3 218,200 55 2.75(±2.13)x10-4 134,400 12 1.00(±0.322)x10-4 2.75 phoboLH312RH236 3 180,900 17 9.05(±3.72)x10-5 175,800 45 2.55(±2.49)x10-4 0.355 aEstimated number of deletion plasmids that were recovered as Kan and Amp resistant colonies. bEstimated number of internal control plasmids that were recovered as Tet and Gent resistant colonies. cTransposition frequency= no. of transposition products/no. of donor plasmids recovered. Values represent the average frequencies from 3 independent injection experiments. Errors are the standard errors of the mean. dRatio of transposition frequency= transposition frequency of the deletion element/transposition frequency of the internal control.
Supplementary Table 2. Transposition frequencies recovered with the full length hobo transposase and an N-terminally truncated form. Helper Plasmids No. of Donor Total No. of Transposition Plasmids Screeneda Transpositions Frequency (x10-4)b
1 pHSH2 333,600 55 1.74±0.283 pKhsphobo 331,200 84 2.66±1.43 pKhsp70hoboATG3 342,600 441 14.2±3.00 pKhsp82hoboATG1 477,800 32 0.696±0.178 pKhsp82hoboATG2 252,200 13 0.580±0.251 pKhsp82hoboATG3 192,200 68 3.48±0.113 a Total no. of donor plasmids recovered from three independent experiments. bAverage value from three independent experiments. Errors are the standard errors of the mean.
Supplementary Table 3. Integration sites of hobo donor elements in target plasmid pGDV1. hoboLH141 hoboLH312 hoboLH567 Site Nuc ± # Site Nuc ± # Site Nuc ± # GATTTGAG 567 + 1 ATAGACGT 429 + 1 GTTTTCTT 605 - 1 GTCTGAAC 736 + 1 ATATGCGT 438 + 1 GTCTGAAC 736 + 3 2 GTACCGAG 2010 + 1 GTCTGAAC 736 + 4 GTTCAGAC 736 - 2 GATTGGAC 830 + 1 ATACAAGC 931 + 1 GTATGCAC 2154 + 1 GTGCATAC 2154 - 2 TATAGAAC 2163 + 1 GTACCAAC 2271 + 2 ATTCAGAG 2358 + 1 GTAAGTAT 2390 + 2
hoboRH65 hoboRH126 hoboRH235 hoboRH682 Site Nuc ± # Site Nuc ± # Site Nuc ± # Site Nuc ± # GTTGGGAT 423 + 2 GTTGGGAT 423 + 1 GTAGTAAT 576 - 1 ATATAAAC 389 - 1 TTTCAAGT 907 - 2 TATAGAAC 2163 + 1 GTTTCTGG 638 - 1 ATCCCAAC 423 - 2 GTATGCAC 2154 + 1 GTTCCGAC 2303 + 4 ATAAATTC 684 - 1 GTCTGAAC 736 + 1 GTACCAAC 2271 + 1 ATACTTAC 2390 - 2 GTCTGAAC 736 + 1 GTTCAGAC 736 - 3 GTTGGTAC 2271 - 1 GGTTTCAA 821 - 1 GTATGCAC 2154 + 1 GTATGCAC 2154 + 1 GTGCATAC 2154 - 4 GTCGGAAC 2303 - 1 GGATTTAA 2254 + 1 TCTTCAAC 2344 + 1 GTTGGTAC 2271 - 1 GTATGTAC 2394 + 1 GTCGGAAC 2303 - 1
hoboRH47512IIR hoboLH312RH235 hobo internal control Site Nuc ± # Site Nuc ± # Site Nuc ± # ATTGAGAT 94 + 1 ATGGTAAG 630 + 1 ATGCAAAT 135 + 1 TAAGGAGT 102 + 1 CTTTGCCA 2031 + 1 GTTTATAT 389 + 1 AATTTATA 685 + 2 GTATGCAC 2154 + 1 TTTACGAG 530 + 1 GTCTGAAC 736 + 2 GTGCATAC 2154 - 1 GTCTGAAC 736 + 4 ATTGAAAA 766 + 1 CAACCAAT 2194 - 1 GTTCAGAC 736 - 1 GTTTTGAT 1931 + 1 GTACATAC 2394 - 2 AAGAGATA 772 + 1 GTATGCAC 2154 + 2 GTACCGAG 2010 + 1 GTGCATAC 2154 - 4 ATTCAGAA 2100 + 1 GTTCCGAC 2303 + 1 CTTTGCCA 2031 + 1 GTCGGAAC 2303 - 1 GTATGCAC 2154 + 10 GTGCATAC 2154 - 2 GTACCAAC 2271 + 8 GTTGGTAC 2271 - 5 GTTCCGAC 2303 + 5 GTCGGAAC 2303 - 1 TCTTCAAC 2344 - 1 GTATGTAC 2394 + 1 GTTAAAAC 2416 - 1 Site refers to the 8 bp direct duplication generated during insertion of hobo donor elements. Nuc refers to the position of insertion in pGDV1. ± refers to the orientation of the integrated hobo element. The (+) orientation is defined for hobo elements inserted into pGDV1 in the 5’ to 3’ direction and the (-) orientation in the 3’ to 5’direction.
3 Supplementary Table 4. Preferred integration sites a of hobo donor elements in pGDV1 Nuc ± Site # 2154 + GTATGCAC 17 736 + GTCTGAAC 16 2154 - GTGCATAC 13 2271 + GTACCAAC 11 2303 + GTCGGAAC 10 2271 - GTTGGTAC 7 736 - GTTCAGAC 6 2303 - GTCGGAAC 4 423 + GTTGGGAT 3 a Sites that have been used three or more times. Site, Nuc, #, and ± are described in Supplementary Table 3. .
4 Supplementary Table 5. Frequencies of pGDV1 insertion sites used by hobo and Hermes. hobo Hermes Nuc Site # of Frequencya # of Frequencyb Insertion(s Insertion(s) ) 2154+ GTATGCAC 17 0.127 14 0.110 736+ GTCTGAAC 16 0.119 12 0.095 2154- GTGCATAC 13 0.097 2 0.016 2271+ GTACCAAC 11 0.082 4 0.032 2303+ GTTCCGAC 10 0.075 10 0.079 2303- GTCGGAAC 4 0.030 2 0.016 423+ GTTGGGAT 3 0.022 4 0.032 2394- GTACATAC 2 0.015 1 0.007 2390+ GTAAGTAT 2 0.015 1 0.007 2010+ GTACCGAG 2 0.015 2 0.016 2100 ATTCAGAA 1 0.007 1 0.007 576- GTAGTAAT 1 0.007 1 0.007 429+ ATAGACGT 1 0.007 3 0.024 389+ GTTTATAT 1 0.007 1 0.007 aFrequency of insertion =number of Hermes insertions for a particular site in pGDV1/ total number of transposition events . The total number of transposition events for Hermes obtained previously by Sarkar et al. (1997a) was 127. bFrequency of insertion =number hobo insertions for a particular site in pGDV1/total number of transposition events. The total number of transposition events for hobo sequenced in this study was 134.
5 6 Supplementary Table 6. Insertion sites in pGDV1 obtained with hsp70 and hsp82 hobo helper plasmidsa. pHSH2 (ORF0-ORF1) pKhsp70hobo (ORF1) pKhsp70hoboATG3 (truncated ORF1) Site Nuc ± # Site Nuc ± # Site Nuc ± # GTCTGAAC 736 + 1 GTTTATAT 389 + 2 AATTTTTC 176 + 1 GTATGGGA 2070 + 1 AGACGTAA 431 + 1 CTTTAGCT 367 - 1 GTGCATAC 2154 - 2 GTCTGAAC 736 + 3 GTTGGGAT 423 + 1 GTTCCGAC 2303 + 1 TTCTACAC 753 - 1 GTCTGAAC 736 + 12 GTCGGAAC 2303 - 1 GTACCGAG 2010 + 1 GTTTATTT 869 + 2 ATTCAGAA 2100 + 1 GTGGCAAA 2032 - 1 GTATGCAC 2154 + 1 ATAAAGGG 2063 + 1 TTCTTCAA 2345 - 1 GTATGCAC 2154 + 4 GTCGGAAC 2303 - 2 GTACATAC 2394 - 2 CTTGGTGT 2401 + 1 GTTTTAGT 2407 + 1 pKhsp82hoboATG1 pKhsp82hoboATG2 pKhsp82hoboATG3 (ORF0-ORF1) (ORF1) (truncated ORF1) Site Nuc ± # Site Nuc ± # Site Nuc ± # GTGTAGAA 753 + 2 GTGTAGAA 753 + 1 TTTGGGAT 339 + 1 GTACATAC 2394 - 1 TGATTGCT 796 + 1 GTATGCAC 2154 + 6 aThe helper plasmids contain either a full-length or truncated transposase reading frame. For depiction of the coding sequences present in these plasmids, see Figure 4. Site, Nuc, #, and ± are explained in Supplementary Table 3.
7