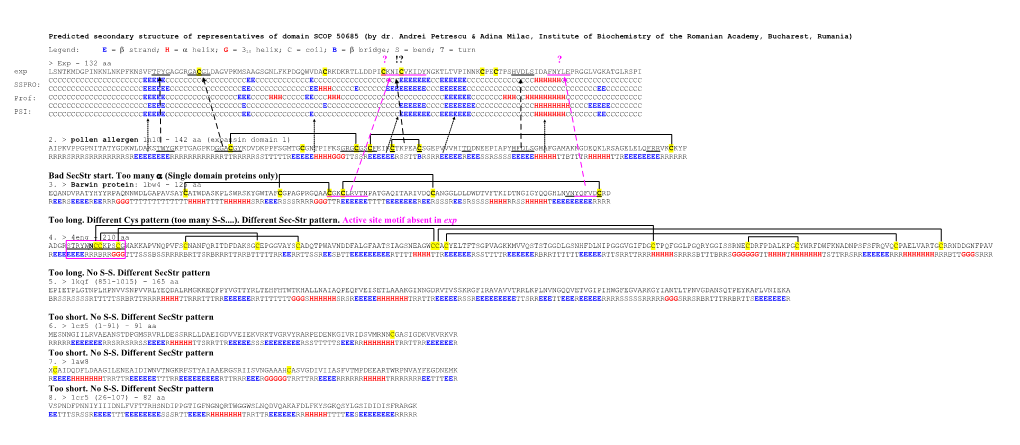
Predicted secondary structure of representatives of domain SCOP 50685 (by dr. Andrei Petrescu & Adina Milac, Institute of Biochemistry of the Romanian Academy, Bucharest, Rumania)
Legend: E = strand; H = helix; G = 310 helix; C = coil; B = bridge; S = bend; T = turn > Exp - 132 aa ? !? ? exp LSNTKMDGPINKNLNKPFKNSVFTFYGAGGR GA CGLDAGVPKMSAAGSGNLFKPDGQWVDACRKDKRTLLDDPI C KNI C VKIDY NGKTLTVPINNKCPECTPSHVDLSIDAFNYLEPRGGLVGKATGLRSPI CCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCCCCCCCCCEECCCCCCCCCCCCECCCCCCCCCCCCCCCCCCCEEEEEEECCEEEEEECCCCCCCCCCCCCCCHHHHHHCCCCCCCCCCCCCCCCCC SSPRO: CCCCCCCCCCCCCCCCCCCCCEEEEEECCCCCCCCCCCCCCCCEECCCCCCCCCCCCCEEHHHCCCCCECCCCCCEEEEEEEEECCCEEEEEECCCCCCCCCCCCCCCCCCCCCCCCCCCCCEECCCCCCCC Prof: CCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCCCCCCCEEECCCCHHHCCCCCEECCCHHHCCCCCCCCCCCCCCCCCCCCCCCEEEEEECCCCCCCHHHCCHHHHHHHHHCCCCCCCCCCCCCCCCC CCCCCCCCCCCCCCCCCCCCCCEEEECCCCCCCCCCCCCCCCCECCCCCCCCCCCCCCCCCECCCCCCCCCCCCCCCCEEEEEECCCCEEEEEEECCCCCCCCCCCCCHHHHHHHHCCCCEEEEECCCCCCC PSI: CCCCCCCCCCCCCCCCCCCCCEEEEECCCCCCCCCCCCCCCCCEECCCCCCCCCCCCCECCCCCCCCCCCCCCCCCCCEEEEEEECCEEEEEEECCCCCCCCCCCCCCHHHHHHHCCCCCCCEECCCCCCCC
2. > pollen allergen 1n10 - 142 aa (expansin domain 1) AIPKVPPGPNITATYGDKWLDAKSTWYGKPTGAGPKD GGA CGYKDVDKPPFSGMTGCGNTPIFKS GRG C GS CFEIKCTKPEACSGEPVVVHITDDNEEPIAPYHFDLSGHAFGAMAKKGDEQKLRSAGELELQFRRVKCKYP RRRRSRRRSRRRRRRRRSREEEEEEEERRRRRRRRRRRRTTRRRRRSSTTTTTREEEEEHHHHGGGTTSSREEEEEERSSTTBRSRREEEEEREEESSRSSSSEEEEEHHHHHTTBTTTRHHHHHTTREEEEEEEERRRRRR
Bad SecStr start. Too many (Single domain proteins only) 3. > Barwin protein: 1bw4 - 125 aa EQANDVRATYHYYRPAQNNWDLGAPAVSAYCATWDASKPLSWRSKYGWTAFCGPAGPRGQAA C GK C LRVTN PATGAQITARIVDQCANGGLDLDWDTVFTKIDTNGIGYQQGHLN VN YQFVD CRD REERSEEEEREERRRGGGTTTTTTTTTTTTTHHHHTTTTHHHHHHSRREEERSSSRRRRGGGTTREEEEEETTTTEEEEEEEREERSSSREESRSSSSHHHHRRSSHHHHHTEEEEEEEEERRRR
Too long. Different Cys pattern (too many S-S....). Different Sec-Str pattern. Active site motif absent in exp
4. > 4eng - 210 aa ADGR STRYW N CC KPS CGWAKKAPVNQPVFSCNANFQRITDFDAKSGCEPGGVAYSCADQTPWAVNDDFALGFAATSIAGSNEAGWCCACYELTFTSGPVAGKKMVVQSTSTGGDLGSNHFDLNIPGGGVGIFDGCTPQFGGLPGQRYGGISSRNECDRFPDALKPGCYWRFDWFKNADNPSFSFRQVQCPAELVARTGCRRNDDGNFPAV REEEEEEERRRBRRGGGTTTSSSBSSRRRRBRTTSRBRRRTTRRBTTTTTRREERRTTSSREESBTTEEEEEEEEERTTTTHHHHTTREEEEEERSSTTTTREEEEEERBRRTTTTTTEEEEERTTSRRTTRRRHHHHHSRRRSBTTTBRRSGGGGGGTTHHHHTHHHHHHHTSTTRRSRREEEEEERRRHHHHHHHRRRBTTGGGSRRR
Too long. No S-S. Different SecStr pattern 5. > 1kqf (851-1015) - 165 aa EPIETPLGTNPLHPNVVSNPVVRLYEQDALRMGKKEQFPYVGTTYRLTEHFHTWTKHALLNAIAQPEQFVEISETLAAAKGINNGDRVTVSSKRGFIRAVAVVTRRLKPLNVNGQQVETVGIPIHWGFEGVARKGYIANTLTPNVGDANSQTPEYKAFLVNIEKA BRSSRSSSSRTTTTTSRBRTTRRRRHHHHTTRRRTTTRREEEEEERRTTTTTTTGGGSHHHHHHSRSREEEEEHHHHHHHTRRTTREEEEERSSREEEEEEEEETTSRREEETTEEEREEEEERRRRSSSSSRRRRRGGGSRRSRBRTTTRRBRTTSEEEEEEER
Too short. No S-S. Different SecStr pattern 6. > 1cz5 (1-91) - 91 aa MESNNGIILRVAEANSTDPGMSRVRLDESSRRLLDAEIGDVVEIEKVRKTVGRVYRARPEDENKGIVRIDSVMRNNCGASIGDKVKVRKVR RRRRREEEEEEERRSRRSRRSSEEEERHHHHHTTSRRTTREEEEESSSEEEEEEEERSSTTTTTSEEERRHHHHHHHTRRTTRREEEEEER Too short. No S-S. Different SecStr pattern 7. > 1aw8 XCAIDQDFLDAAGILENEAIDIWNVTNGKRFSTYAIAAERGSRIISVNGAAAHCASVGDIVIIASFVTMPDEEARTWRPNVAYFEGDNEMK REEEEHHHHHHHTRRTTREEEEEETTTRREEEEEEEEERTTRRREEERGGGGGTRRTTRREEEERRRRRRHHHHHTRRRRRRREETTTEER Too short. No S-S. Different SecStr pattern 8. > 1cr5 (26-107) - 82 aa VSPNDFPNNIYIIIDNLFVFTTRHSNDIPPGTIGFNGNQRTWGGWSLNQDVQAKAFDLFKYSGKQSYLGSIDIDISFRARGK EETTTSRSSREEEETTTEEEEEEEESSSRTTEEEERHHHHHHHTRRTTREEEEEERRHHHHHTTTTEESEEEEEEEERRRRR SCOP Fold classification: Double psi beta-barrel
Superfamilies: 1. Barwin-like endoglucanases: Families: Endoglucanase V (Eng V): 4eng Pollen allergen PHL P 1 N-terminal domain (1003-1145): 1n10 Barwin (basic barley seed protein - lectin): 1bw4 2. ADC-like Families: Pyruvoyl dependent aspartate decarboxylase, ADC 1aw8 Formate dehydrogenase/DMSO reductase, C-terminal domain 1kqf (851-1015) Cdc48 N-terminal domain-like 1cr5 (A26-A107), 1cz5 (1-91), 1e32 (21-106)
DALI fold classification: DC_A_B_C_D (A= the class; B=the globular folding topology; C=the functional family; D=the sequence family) 1n10 - does not appear as a representative (separate entry) in DALI classification. By coordinate search 1n10 DC_6_107 6: class 6 - domains which are not clearly closer to one of the 5 main attractors (1 - alpha/beta; 2 - all-beta; 3 - all-alpha; 4 - antiparallel beta-barrels; 5 - alpha-beta meander) 107: the cluster number of structural neighbours in fold space with an average Dali pairwise Z-score > 2)
1: 7585-A 1n10-A 25.7 0.0 133 228 100 0 0 1 S pollen allergen phl p 1 (phl p i) 2: 7585-A 1bw4 7.0 2.9 97 125 18 0 0 14 S Barwin, basic barley seed protein DC_6_107_5_1 3: 7585-A 1cz4-A 6.8 2.6 86 185 7 0 0 10 S vcp-like atpase fragment DC_6_107_2_3 [4: 7585-A 1e32-A 6.7 2.6 81 438 5 0 0 8 S p97 fragment] 5: 7585-A 2eng 6.5 2.5 90 205 26 0 0 10 S endoglucanase v DC_6_107_4_1 6: 7585-A 1qcs-A 6.4 2.5 78 195 5 0 0 8 S n-ethylmaleimide sensitive factor (nsf-n) fragment DC_6_107_2_2 7: 7585-A 1cr5-A 6.0 2.3 73 178 11 0 0 8 S sec18p (residues 22 - 210) fragment DC_6_107_2_1 8: 7585-A 1dmr 5.0 9.4 93 779 2 0 0 11 S dmso reductase biological_unit DC_6_107_1_2 [9: 7585-A 1kqf-A 4.8 2.9 79 982 6 0 0 8 S formate dehydrogenase, nitrate-inducible, major subunit] CATH fold classification: 1n10: does not appear yet in CATH as a separate entry (Found 0 domains and 2 preliminary data entries: 1n10A & 1n10B). Class 2: Mainly Beta Architecture: 2.40 Barrel Topology: 2.40.40 - Barwin-like endoglucanases Homologous Superfamily: 2.40.40.10 - LECTIN 2.40.40.10.1 - 1bw300 - LECTIN 2.40.40.10.2 - 2eng00 - HYDROLASE (ENDOGLUCANASE) 2.40.40.20 - 1eu1A4 - OXIDOREDUCTASE 2.40.40.20.1 - 1eu1A4 - OXIDOREDUCTASE 2.40.40.20.2 - 1aw8B0 - DECARBOXYLASE 2.40.40.20.3 - 2napA4 - OXIDOREDUCTASE 2.40.40.20.4 - 1cr5A1 - ENDOCYTOSIS/EXOCYTOSIS 2.40.40.20.5 - 1cz4A1 - HYDROLASE 2.40.40.20.6 - 1qcsA1 - FUSION PROTEIN 2.40.40.20.7 - 1e32A1 - ATPASE 2.40.40.20.8 - 1g8kA3 - OXIDOREDUCTASE 2.40.40.30 - 1i50A3 - TRANSCRIPTION 2.40.40.30.1 - 1i50A3 - TRANSCRIPTION