Supplementary material:
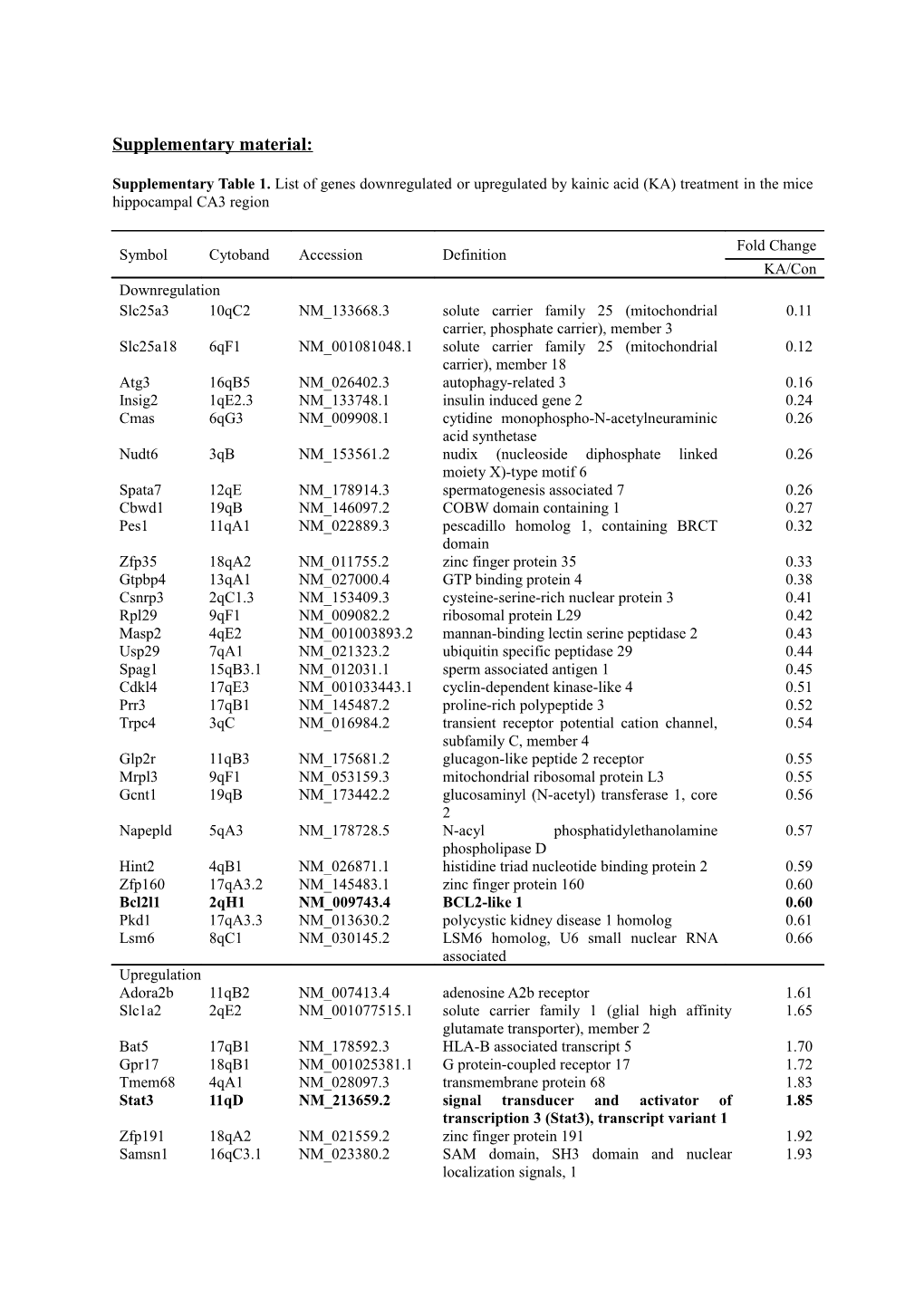
Supplementary Table 1. List of genes downregulated or upregulated by kainic acid (KA) treatment in the mice hippocampal CA3 region
Fold Change Symbol Cytoband Accession Definition KA/Con Downregulation Slc25a3 10qC2 NM_133668.3 solute carrier family 25 (mitochondrial 0.11 carrier, phosphate carrier), member 3 Slc25a18 6qF1 NM_001081048.1 solute carrier family 25 (mitochondrial 0.12 carrier), member 18 Atg3 16qB5 NM_026402.3 autophagy-related 3 0.16 Insig2 1qE2.3 NM_133748.1 insulin induced gene 2 0.24 Cmas 6qG3 NM_009908.1 cytidine monophospho-N-acetylneuraminic 0.26 acid synthetase Nudt6 3qB NM_153561.2 nudix (nucleoside diphosphate linked 0.26 moiety X)-type motif 6 Spata7 12qE NM_178914.3 spermatogenesis associated 7 0.26 Cbwd1 19qB NM_146097.2 COBW domain containing 1 0.27 Pes1 11qA1 NM_022889.3 pescadillo homolog 1, containing BRCT 0.32 domain Zfp35 18qA2 NM_011755.2 zinc finger protein 35 0.33 Gtpbp4 13qA1 NM_027000.4 GTP binding protein 4 0.38 Csnrp3 2qC1.3 NM_153409.3 cysteine-serine-rich nuclear protein 3 0.41 Rpl29 9qF1 NM_009082.2 ribosomal protein L29 0.42 Masp2 4qE2 NM_001003893.2 mannan-binding lectin serine peptidase 2 0.43 Usp29 7qA1 NM_021323.2 ubiquitin specific peptidase 29 0.44 Spag1 15qB3.1 NM_012031.1 sperm associated antigen 1 0.45 Cdkl4 17qE3 NM_001033443.1 cyclin-dependent kinase-like 4 0.51 Prr3 17qB1 NM_145487.2 proline-rich polypeptide 3 0.52 Trpc4 3qC NM_016984.2 transient receptor potential cation channel, 0.54 subfamily C, member 4 Glp2r 11qB3 NM_175681.2 glucagon-like peptide 2 receptor 0.55 Mrpl3 9qF1 NM_053159.3 mitochondrial ribosomal protein L3 0.55 Gcnt1 19qB NM_173442.2 glucosaminyl (N-acetyl) transferase 1, core 0.56 2 Napepld 5qA3 NM_178728.5 N-acyl phosphatidylethanolamine 0.57 phospholipase D Hint2 4qB1 NM_026871.1 histidine triad nucleotide binding protein 2 0.59 Zfp160 17qA3.2 NM_145483.1 zinc finger protein 160 0.60 Bcl2l1 2qH1 NM_009743.4 BCL2-like 1 0.60 Pkd1 17qA3.3 NM_013630.2 polycystic kidney disease 1 homolog 0.61 Lsm6 8qC1 NM_030145.2 LSM6 homolog, U6 small nuclear RNA 0.66 associated Upregulation Adora2b 11qB2 NM_007413.4 adenosine A2b receptor 1.61 Slc1a2 2qE2 NM_001077515.1 solute carrier family 1 (glial high affinity 1.65 glutamate transporter), member 2 Bat5 17qB1 NM_178592.3 HLA-B associated transcript 5 1.70 Gpr17 18qB1 NM_001025381.1 G protein-coupled receptor 17 1.72 Tmem68 4qA1 NM_028097.3 transmembrane protein 68 1.83 Stat3 11qD NM_213659.2 signal transducer and activator of 1.85 transcription 3 (Stat3), transcript variant 1 Zfp191 18qA2 NM_021559.2 zinc finger protein 191 1.92 Samsn1 16qC3.1 NM_023380.2 SAM domain, SH3 domain and nuclear 1.93 localization signals, 1 Klc1 12qF1 NM_001025360.2 kinesin light chain 1 (Klc1), transcript variant 2.03 d Eif1b 9qF4 NM_026892.1 eukaryotic translation initiation factor 1B 2.04 Arl5a 2qC1.1 NM_182994.2 ADP-ribosylation factor-like 5A 2.10 Ppp1r13b 12qF1 NM_011625.1 protein phosphatase 1, regulatory (inhibitor) 2.15 subunit 13B H2-T10 17qB1 NM_010395.5 histocompatibility 2, T region locus 10 2.18 Slc35f4 14qC1 NM_029238.2 solute carrier family 35, member F4 2.18 Mrps12 7qA3 NM_011885.3 mitochondrial ribosomal protein S12 2.19 Ypel4 2qD NM_001005342.2 yippee-like 4 2.20 Smarcc1 9qF2 NM_009211.2 SWI/SNF related, matrix associated, actin 2.20 dependent regulator of chromatin, subfamily c, member 1 Acad9 3qB NM_172678.3 acyl-Coenzyme A dehydrogenase family, 2.25 member 9 Usp38 8qC2 NM_027554.2 ubiquitin specific peptidase 38 2.29 Abhd6 14qA1 NM_025341.3 abhydrolase domain containing 6 2.31 Nt5dc3 10qC1 NM_175331.3 5-nucleotidase domain containing 3 2.39 Chst10 1qB NM_145142.2 carbohydrate sulfotransferase 10 2.47 Timp1 XqA1.3 NM_011593.2 tissue inhibitor of metalloproteinase 1 2.51 Pop4 7qB2 NM_025390.4 processing of precursor 4, ribonuclease 2.55 P/MRP family, Axud1 9qF4 NM_153287.3 AXIN1 up-regulated 1 2.56 Ly6a 15qD3 NM_010738.2 lymphocyte antigen 6 complex, locus A 2.61 Neu1 17qB1 NM_010893.2 neuraminidase 1 2.63 Glo1 17qA3.3 NM_025374.2 glyoxalase 1 2.89 Nup133 8qE2 NM_172288.1 nucleoporin 133 3.12 Slc7a14 3qA3 NM_172861.2 solute carrier family 7 (cationic amino acid 3.16 transporter, y+ system), member 14 Gfap 11qE1 NM_010277.2 glial fibrillary acidic protein 3.23 Rapgefl1 11qD NM_001080925.1 Rap guanine nucleotide exchange factor 3.60 (GEF)-like 1 Ccr6 17qA1 NM_009835.2 chemokine (C-C motif) receptor 6 4.40 Rbm45 2qC3 NM_153405.2 RNA binding motif protein 45 4.74 Centg1 10qD3 NM_001033263.1 centaurin, gamma 1 4.83 Gdi1 XqA7.3 NM_010273.3 guanosine diphosphate (GDP) dissociation 4.89 inhibitor 1 Tesk1 4qB1 NM_011571.2 testis specific protein kinase 1 5.50 Manbal 2qH1 NM_026968.2 mannosidase, beta A, lysosomal-like 5.55 Psmd8 7qB1 NM_026545.2 proteasome (prosome, macropain) 26S 10.25 subunit, non-ATPase, 8 Csrp1 1qE4 NM_007791.4 cysteine and glycine-rich protein 1 10.47 Spink8 9qF2 NM_183136.2 serine peptidase inhibitor, Kazal type 8 10.93 Kns2 12qF1 NM_001025360.1 kinesin 2 (Kns2), transcript variant d 11.27 Genes that adjusted P-value is < 0.05, and that fold change is 1.5 ≤ or < 0.7 are selected. Adjusted P-value is the adjusted false discovery rate (FDR) P-value of the moderated ANOVA test followed by Tukey’s HSD post-hoc test of the microarray.