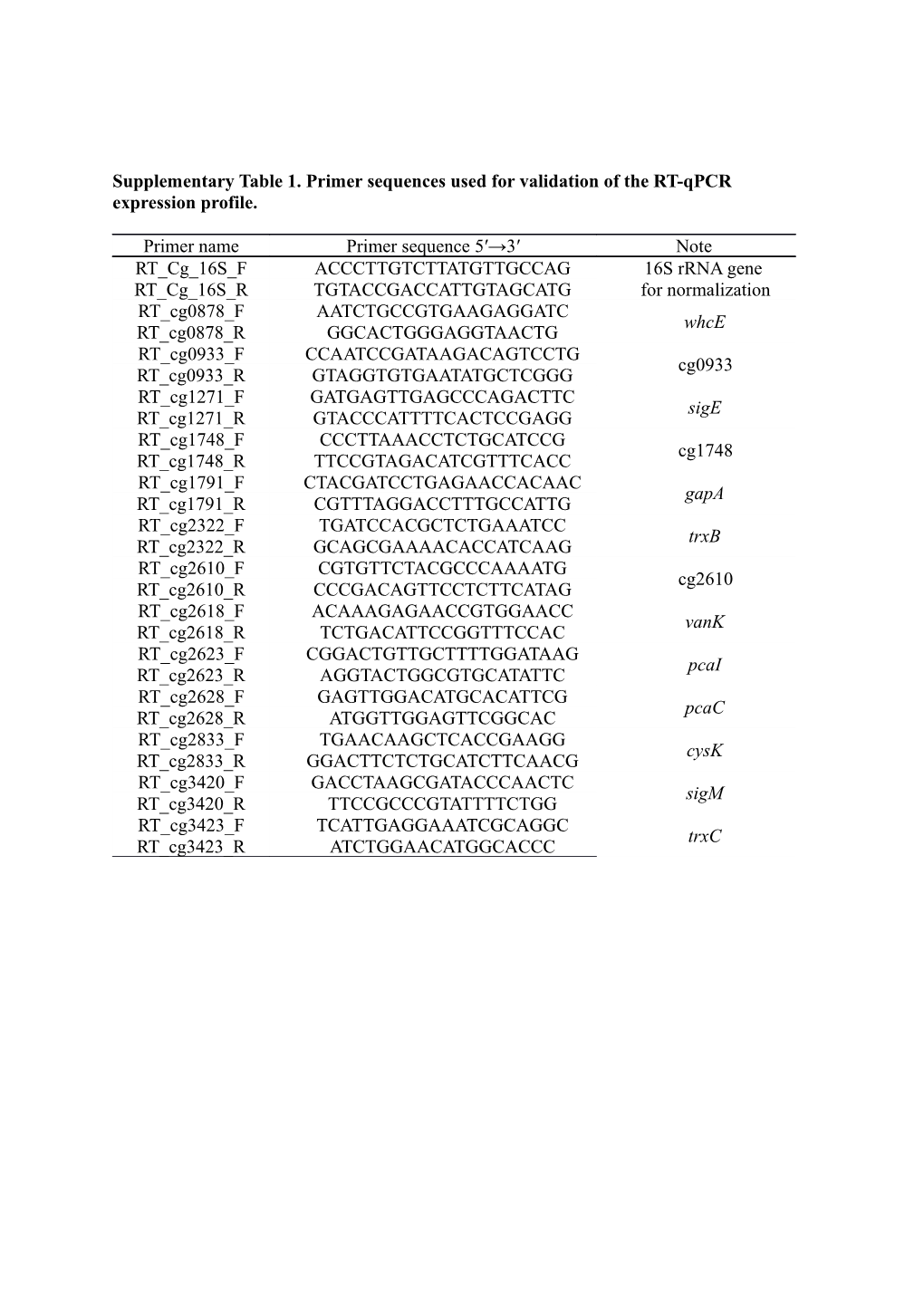
Supplementary Table 1. Primer sequences used for validation of the RT-qPCR expression profile.
Primer name Primer sequence 5′→3′ Note RT_Cg_16S_F ACCCTTGTCTTATGTTGCCAG 16S rRNA gene RT_Cg_16S_R TGTACCGACCATTGTAGCATG for normalization RT_cg0878_F AATCTGCCGTGAAGAGGATC whcE RT_cg0878_R GGCACTGGGAGGTAACTG RT_cg0933_F CCAATCCGATAAGACAGTCCTG cg0933 RT_cg0933_R GTAGGTGTGAATATGCTCGGG RT_cg1271_F GATGAGTTGAGCCCAGACTTC sigE RT_cg1271_R GTACCCATTTTCACTCCGAGG RT_cg1748_F CCCTTAAACCTCTGCATCCG cg1748 RT_cg1748_R TTCCGTAGACATCGTTTCACC RT_cg1791_F CTACGATCCTGAGAACCACAAC gapA RT_cg1791_R CGTTTAGGACCTTTGCCATTG RT_cg2322_F TGATCCACGCTCTGAAATCC trxB RT_cg2322_R GCAGCGAAAACACCATCAAG RT_cg2610_F CGTGTTCTACGCCCAAAATG cg2610 RT_cg2610_R CCCGACAGTTCCTCTTCATAG RT_cg2618_F ACAAAGAGAACCGTGGAACC vanK RT_cg2618_R TCTGACATTCCGGTTTCCAC RT_cg2623_F CGGACTGTTGCTTTTGGATAAG pcaI RT_cg2623_R AGGTACTGGCGTGCATATTC RT_cg2628_F GAGTTGGACATGCACATTCG pcaC RT_cg2628_R ATGGTTGGAGTTCGGCAC RT_cg2833_F TGAACAAGCTCACCGAAGG cysK RT_cg2833_R GGACTTCTCTGCATCTTCAACG RT_cg3420_F GACCTAAGCGATACCCAACTC sigM RT_cg3420_R TTCCGCCCGTATTTTCTGG RT_cg3423_F TCATTGAGGAAATCGCAGGC trxC RT_cg3423_R ATCTGGAACATGGCACCC Supplementary Table 2. Transcriptional regulators based on a transcriptome comparison of Corynebacterium glutamicum ATCC 13032 with the wild-type strain cultivated under the no stress condition and the H2O2-adapted strain cultivated in MCGC containing 10 mM H2O2.
Gene Gene Protein Expression Annotationc IDa nameb family ratiod Carbon metabolism Response Activator of citrate transporter genes citH and cg0090 citB 1.99 regulator tctCBA Dual regulator (global regulator) functioning as cg0350 glxR Crp 0.76 network hub Dual regulator (master regulator) of carbon cg0444 ramB HTH_3 1.55 metabolism Activator of propionate utilization genes cg0800 prpR HTH_3 2.09 prpD2B2C2 Repressor of ribose uptake and uridine cg1410 rbsR LacI 2.12 utilization genes Repressor of uridine utilization and ribose cg1547 uriR LacI 2.22 uptake genes cg1738 acnR TetR Repressor of aconitase gene acn 2.29 cg1935 gntR2 GntR Dual regulator involved in carbon metabolism 1.63 Repressor (master regulator) involved in carbon cg2115 sugR DeoR 2.15 metabolism cg2118 fruR DeoR Repressor of fructose PTS system genes 0.98 cg2746 - COG2508 putative sugar diacid utilization regulator 2.12 cg2783 gntR1 GntR Dual regulator involved in carbon metabolism 1.74 Dual regulator (master regulator) of carbon cg2831 ramA LuxR 1.43 metabolism cg3224 lldR GntR Repressor involved in carbon metabolism 1.74 Amino acid and vitamin biosynthesis and anaerobic respiration cg0313 lrp AsnC Activator of amino acid exporter genes brnFE 2.23 Activator of serine hydroxymethyltransferase cg0527 glyR ArsR 0.43 gene glyA Activator of pyridoxal phosphate synthase cg0897 pdxR GntR 1.18 genes Repressor of NAD biosynthesis genes nadAC- cg1218 nrtR NrtR 0.56 cg1214 Dual regulator (master regulator) of anaerobic cg1340 arnR ArnR 0.95 respiration cg1425 lysG LysR Activator of amino acid exporter gene lysE 2.33 cg1486 ltbR IclR Repressor of leucine and tryptophan 1.74 Repressor of arginine and glutamate cg1585 argR ArgR 2.11 biosynthesis genes Repressor of ribonucleotidereductase genes cg2112 nrdR NrdR 2.47 nrdHIE Repressor of biotin uptake and biosynthesis cg2309 bioQ TetR 1.70 genes cg2894 cgmR TetR Repressor of MFS-type transporter gene 1.50 cg2893 Repressor of arginine and glutamate cg3202 farR GntR 0.92 biosynthesis genes cg3261 - GntR Regulator of conserved hypothetical protein 1.17 Aromatic compound catabolism cg0500 qsuR LysR Activator of quinate/ shikimate pathway genes 1.76 cg1308 rhcR TetR Repressor of resorcinol pathway genes 2.22 cg2615 vanR PadR Repressor of vanillate pathway genes 3.38 cg2624 pcaR IclR Repressor of protocatechuate pathway genes 4.09 cg2627 pcaO LuxR Activator of protocatechuate pathway genes 3.64 cg2641 benR LuxR Activator of catechol pathway genes 3.71 cg2965 - AraC Activator of phenol pathway gene 2.30 cg3352 genR IclR Activator of gentisate pathway genes 2.04 Activator of putative hydroxyquinol pathway cg3388 - IclR 1.91 genes Macroelement metabolism Activator of sulfonate and sulfonate ester cg0012 ssuR ROK 0.91 utilization cg0112 ureR MarR putative repress or urease genes 2.53 Dual regulator of assimilatory sulfate reduction cg0156 cysR ROK 1.33 genes cg0986 amtR TetR Repressor of nitrogen metabolism 1.22 Response cg2888 phoR Activator of phosphate metabolism 1.44 regulator cg3253 mcbR TetR Repressor of sulfur metabolism 1.70 Metal homeostasis Repressor of arsenitepermease and arsenate Cg0317 arsR2 ArsR 1.89 reductase genes Cg1120 ripA AraC Repressor of iron protein genes 0.68 Repressor of arsenitepermease and arsenate Cg1704 arsR1 ArsR 1.34 reductase genes Cg2103 dtxR DtxR Dual regulator of iron metabolism 1.40 Cg2500 znr ArsR Putative repressor of zinc uptake regulator gene 0.81 Cg2502 zur FUR Repressor of zinc uptake system genes 1.08 Sigma factors cg0309 sigC SigC ECF sigma factor of unknown function 1.98 ECF sigma factor of probably involved in the cg0696 sigD SigD 1.46 adaptation to microaerobic environments ECF sigma factor controlling the heat and cg0876 sigH SigH 2.12 oxidative stress response ECF sigma factor involved in responses to cells cg1271 sigE SigE 0.09 surface stresses cg2092 sigA SigA Primary (housekeeping) sigma factor 0.74 Nonessential primary-like sigma factor involved in gene expression during the cg2102 sigB SigB 0.66 transition phase, under oxygen deprivation and during environmental stress responses ECF sigma factor controlling the expression of cg3420 sigM SigM 1.79 disulfide stress-related genes Anti-sigma factors cg0877 rhsA RhsA Anti-sigma factor of SigH 1.80 cg1272 cseE CseE Anti-sigma factor of SigE 1.89 SOS and stress response Repressor of cg0215 cspA Cold malate synthase 0.88 gene aceB Repressor of cg0337 whcA WhiB oxidative stress 1.68 response genes Stationary cg0695 whcB WhiB phase-specific 0.52 regulator Dual regulator of genes involved in cell Response morphology, cg0862 mtrA 1.41 regulator antibiotics susceptibility and osmoprotection Activator of cg0878 whcE WhiB thioredoxin 0.13 genes Hydrogen peroxide- sensitive MarR- cg1324 rosR RosR 1.89 type transcriptional regulator putative stress- responsive cg1325 - - 1.77 transcriptional regulator Repressor of cg1552 qorR COG1733 quinoneoxidored 1.25 uctase gene qor2 Repressor of iron–sulfur cg1765 sufR ArsR 0.63 cluster biogenesis genes Hydrogen cg2109 oxyR LysR peroxide sensing 1.35 regulator Dual regulator (master cg2114 lexA LexA 1.51 regulator) of SOS response cg2152 clgR HTH_3 Activator of 1.62 proteolysis and DNA repair genes Repressor of heat shock cg2516 hrcA HrcA response genes 2.08 with CIRCE elements Repressor of heat shock cg3097 hspR MerA response genes 1.91 with HAIR elements a Gene ID according to the accession number BX927147 was used. b,c Gene name and annotation was obtained form the CoryneRegNet database (www.coryneregnet.de), and Brinkrolf et al., 2010, and Schröderand Tauch, 2010. d Expression ratios are from a comparison of the H2O2-adapted strain under the oxidative stress condition vs. the wild-type strain under the no stress condition. Supplementary Table 3. Oxidation-reduction related genes from the transcriptome results.
Gene Gene Protein Expression Annotationd IDa nameb IDc ratioe cg003 Reductase related to - YP_224316.1 2.62 1 diketogulonatereductase cg006 - YP_224345.1 Haem peroxidase superfamily 2.39 5 cg013 - YP_224397.1 putative oxidoreductase 2.02 1 cg020 putative oxidoreductasemyo-inositol 2- iolG YP_224460.1 2.15 4 dehydrogenase cg020 oixA YP_224463.1 hypothetical oxidoreductase 2.32 7 cg0211 - YP_224467.1 putative oxidoreductase 1.90 cg023 - YP_224488.1 putative oxidoreductase 1.03 7 cg023 - YP_224489.1 L-Gulonolactone oxidase 0.77 8 cg025 - YP_224502.1 Quinone oxidoreductase 1.85 1 cg025 - YP_224504.1 Flavodoxinreductase 1.71 3 cg027 - YP_224523.1 putative oxidoreductase 2.14 4 cg031 katA YP_224555.1 catalase 2.11 0 cg031 arsX YP_224563.1 Arsenate reductase 1.93 9 cg032 nuoL YP_224569.1 NADH-quinoneoxidoreductase chain 5 2.06 6 cg034 fabG1 YP_224585.1 3-Oxoacyl-(acyl-carrier protein) reductase 2.32 4 cg040 dTDP-4-dehydrorhamnose 3,5-epimerase, rmlCD YP_224633.1 1.30 2 dTDP-dehydrorhamnosereductase cg040 - YP_224635.1 Nitroreductase family 0.74 4 cg042 UDP-N- murB YP_224653.1 1.72 3 acetylenolpyruvoylglucosaminereductase cg047 Probable UDP-N- murB2 YP_224698.1 1.00 6 acetylenolpyruvoylglucosamine reductase cg049 proC YP_224712.1 Pyrroline-5-carboxylate reductase 1.32 0 cg049 hemA YP_224719.1 Glutamyl-tRNAreductase 2.14 7 cg051 hemY YP_224738.1 putative protoporphyrinogen oxidase 2.15 7 cg061 dkg YP_224819.1 2,5-diketo-D-gluconic acid reductase 1.42 2 cg061 putative formate dehydrogenase fdhF YP_224823.1 1.39 8 oxidoreductase protein cg062 - YP_224828.1 Secreted oxidoreductase 2.30 4 cg063 putative betaine aldehyde dehydrogenase betB YP_224839.1 1.92 7 (BADH) oxidoreductase cg063 - YP_224841.1 Ferredoxinreductase 2.23 9 cg070 IMP dehydrogenase/ GMP reductase C guaB3 YP_224897.1 1.38 0 terminus cg079 FAD-dependent pyridine nucleotide- - YP_224984.1 2.54 5 disulphideoxidoreductase cg082 - YP_225012.1 putative dihydrofolatereductase 1.55 8 cg096 folA YP_225135.1 Dihydrofolatereductase 1.58 5 cg106 - YP_225228.1 Probable oxidoreductase 1.93 8 cg108 - YP_225238.1 putative multicopper oxidase 1.64 0 Pyrimidine reductase, riboflavin cg1118 - YP_225273.1 2.21 biosynthesis cg1150 - YP_225302.1 NADPH-dependent FMN reductase 2.03 cg1192 - YP_225338.1 Aldo/ketoreductase 2.16 cg123 tpx YP_225377.1 Thiol peroxidase 0.43 6 cg123 - YP_225380.1 Similar to ketopantoatereductase 0.97 9 cg124 Arsenate reductase or related protein, - YP_225386.1 1.53 4 glutaredoxin family cg130 Cytochrome d terminal oxidase cydB YP_225440.1 2.42 0 polypeptide subunit cg130 cydA YP_225441.1 Cytochrome d ubiquinol oxidase subunit I 2.50 1 2-polyprenyl-6-methoxyphenol cg130 rhcH YP_225448.1 hydroxylase and related FAD-dependent 2.42 9 oxidoreductase cg131 rhcM1 YP_225449.1 Maleylacetatereductase 2.32 0 cg134 narI YP_225476.1 Nitrate reductase gamma subunit 1.85 1 cg134 narJ YP_225477.1 Nitrate reductase delta subunit 2.20 2 cg134 narH YP_225478.1 Nitrate reductase beta subunit 2.12 3 cg134 narG YP_225479.1 Nitrate reductase alpha subunit 2.30 4 cg142 putative oxidoreductase (related to aryl- - YP_225550.1 1.67 3 alcohol dehydrogenases) cg152 putative 2,5-diketo-D-gluconic acid dkgX YP_225642.1 2.16 8 reductase cg158 N-acetyl-gamma-glutamyl-phosphate argC YP_225681.1 2.11 0 reductase cg168 - YP_225770.1 Short-chain dehydrogenase/reductase 2.30 0 cg170 FAD-dependent pyridine nucleotide- - YP_225793.1 1.81 3 disulphideoxidoreductase cg1711 - YP_225801.1 Oxidoreductase 2.29 cg176 ctaA YP_225854.1 Cytochrome oxidase assembly protein 1.05 9 cg177 Probable NADPH: quinonereductase, qor YP_225856.1 1.83 1 zeta-crystallin cg177 Polyprenyltransferase (cytochrome ctaB YP_225857.1 2.14 3 oxidase assembly factor) cg178 'soxA YP_225864.1 Sarcosine oxidase-fragment 2.20 1 cg178 soxA YP_225865.1 Sarcosine oxidase-N-terminal fragment 2.46 3 cg180 putative bifunctional riboflavin-specific ribG YP_225881.1 2.05 0 deaminase/reductase Coenzyme F420-dependent N5,N10- cg184 methylene - YP_225927.1 1.54 8 tetrahydromethanopterinreductase or related flavin-dependent cg188 Predicted iron-dependent peroxidase, - YP_225955.1 2.13 1 secreted protein cg207 Peptide methionine sulfoxidereductase- msrB YP_226139.1 0.96 8 related protein cg216 dapB YP_226215.1 Dihydrodipicolinatereductase 2.27 3 cg219 mqo YP_226243.1 Malate: quinoneoxidoreductase 1.59 2 cg219 gor/mtr YP_226245.1 putative glutathione reductase 2.35 4 cg223 putative D-amino acid oxidase/ thiO YP_226279.1 2.30 7 flavoproteinoxidoreductase Coenzyme F420-dependent N5,N10- cg232 - YP_226360.1 methylene 2.21 9 tetrahydromethanopterinreductase cg238 metF YP_226413.1 5,10-methylenetetrahydrofolate reductase 2.20 3 cg240 ctaE YP_226434.1 Cytochrome c oxidase subunit 3 2.30 6 cg240 ctaC YP_226437.1 Cytochrome c oxidase chain II 1.71 9 cg251 hemN YP_226535.1 putative anaerobic coproporphyrinogen III 1.72 7 oxidase cg254 Probable glycolate oxidase (FAD-linked glcD YP_226558.1 2.23 3 subunit) oxidoreductase cg258 proA YP_226599.1 Gamma-glutamyl phosphate reductase 1.14 6 cg259 dkgA YP_226604.1 2,5-diketo-d-gluconic acid reductase 1.42 1 cg261 Malate dehydrogenase oxidoreductase mdh YP_226625.1 1.62 3 protein cg263 benC YP_226650.1 Benzoate dioxygenasereductase 4.55 9 cg267 AlkylhydroperoxidaseAhpD family core - YP_226677.1 0.82 4 domain cg268 putative short-chain - YP_226686.1 0.90 5 dehydrogenase/reductase cg273 Probable bacterioferritin comigratory bcp YP_226730.1 1.68 6 oxidoreductase cg278 Probable cytochrome c oxidase ctaD YP_226765.1 1.02 0 polypeptide subunit cg278 Ribonucleoside-diphosphatereductase 2 nrdF YP_226766.1 1.63 1 beta chain cg278 Ribonucleoside-diphosphatereductase nrdE YP_226771.1 1.32 6 alpha chain cg279 NADPH quinonereductase or related Zn- - YP_226778.1 1.91 5 dependent oxidoreductase cg286 gpx YP_226832.1 Glutathione peroxidase 2.24 7 cg291 - YP_226877.1 putative oxidoreductase 2.56 9 cg295 L-2,3-Butanediol butA YP_226913.1 0.22 8 dehydrogenase/acetoinreductase 2-Polyprenyl-6-methoxyphenol cg296 - YP_226920.1 hydroxylase or related FAD-dependent 2.25 6 oxidoreductase cg299 - YP_226947.1 putative ferredoxinreductase 1.86 9 cg300 Probable succinate-semialdehyde gabD1 YP_226951.1 2.23 4 dehydrogenase oxidoreductase NADH ubiquinone oxidoreductase cg302 mrpA YP_226967.1 subunit 5 (chain L)/Multisubunit 1.96 4 Na+/H+antiporter, A subunit NADH-ubiquinone cg302 mrpD YP_226969.1 oxidoreductase/Multisubunit 1.96 6 Na+/H+antiporter, D subunit cg304 putative ferredoxin/ferredoxin-NADP fprA YP_226992.1 1.68 9 reductase cg309 2-polyprenylphenol hydroxylase or related - YP_227029.1 1.47 2 flavodoxinoxidoreductase cg3116 cysH YP_227054.1 Phosphoadenosine- 0.60 phosphosulfatereductase cg3118 cysI YP_227056.1 Sulfite reductase (hemoprotein) 0.83 cg3119 cysJ YP_227057.1 Probable sulfite reductase (flavoprotein) 1.01 cg313 - YP_227073.1 Nitroreductase 1.68 6 cg322 ldh YP_227152.1 NADPH-dependent FMN reductase 1.48 3 cg323 msrA YP_227165.1 Peptide methionine sulfoxidereductase 2.01 6 cg328 copO Secreted multicopper oxidase 1.76 7 cg329 Probable oxidoreductase protein; - YP_227216.1 1.77 0 NADPH: quinonereductase cg333 - YP_227255.1 putative quinoneoxidoreductase 1.94 2 putative FAD-dependent pyridine cg333 merA YP_227261.1 nucleotide-disulphideoxidoreductase, 1.89 9 similar to mercuric reductases cg334 - YP_227266.1 Nitroreductase 2.26 4 cg335 Probable gentisate 1,2-dioxygenase genD YP_227273.1 2.15 1 oxidoreductase cg337 putative NADH-dependent - YP_227291.1 2.18 0 flavinoxidoreductase cg337 putative NDAH-dependent cye1 YP_227295.1 2.34 4 flavinoxidoreductase cg338 - YP_227300.1 putative oxidoreductase protein 2.13 0 cg338 rhcM2 YP_227305.1 Maleylacetatereductase 2.17 6 cg340 NADPH quinonereductase or Zn- - YP_227324.1 1.61 5 dependent oxidoreductase cg342 trxB YP_227339.1 Thioredoxinreductase 1.98 2 a Gene ID according to the accession number BX927147 was used. b, c, d Gene name, protein ID, and annotation were obtained from the CoryneRegNet database (www.coryneregnet.de) and Brinkrolf et al., 2010 and Schröder and Tauch, 2010. e Expression ratios from a comparison of the H2O2-adapted strain under the oxidative stress condition vs. the wild-type strain under the no stress condition. Supplementary Table 4. List of genes showing a ≥ 3-fold increase in comparison with the H2O2-adapted and wild-type strains.
Gene Gene Expression No Annotationc IDa nameb ratiod 1 cg0311 - Secreted protein 3.12 2 cg1328 - putative Copper chaperone 3.06 3 cg1748 - putative secreted protein 3.40 4 cg1912 - hypothetical protein 3.22 5 cg1925 - hypothetical protein 3.40 6 cg1932 ppp2 Probable protein phosphatase 3.02 7 cg1938 - hypothetical protein 3.23 8 cg1974 - putative lysin 3.20 9 cg2003 - hypothetical protein 3.03 10 cg2156 - hypothetical protein 3.25 11 cg2600 tnp1d Transposase (ISCg1d) 3.50 12 cg2608 folC Folylpolyglutamate synthase 3.98 13 cg2609 valS putative valine-tRNA ligase 3.29 ABC-type dipeptide/oligopeptide/nickel transport cg2610 - 4.20 14 system, secreted component 15 cg2614 - Bacterial regulatory proteins, TetR family 3.51 16 cg2615 vanR Putative transcriptional regulator PadR-like family 3.38 17 cg2616 vanA Probable vanillateO-demethylaseoxygenase subunit 3.89 18 cg2617 vanB Probable VanillateO-demethylase 3.39 19 cg2618 vanK Vanillate transporter Vank 4.11 20 cg2619 - Predicted permease 3.96 Probable succinyl-CoA:3-ketoacid-coenzyme A 21 cg2622 pcaJ transferase subunit acyl-CoA: acetate CoA 4.35 transferase beta subunit Probable fesuccinyl-CoA:3-ketoacid-coenzyme A 22 cg2623 pcaI transferase subunit or acyl-CoA:acetate CoA 4.45 transferase alpha subunit Repressor of protocatechuate pathway genes. IclR cg2624 pcaR 4.09 23 family 24 cg2625 pcaF Putative Acetyl-CoA:Acetyltransferase 4.35 3-Oxoadipate enol-lactone hydrolase/4- cg2626 pcaD 3.35 25 Carboxymuconolactonedecarboxylase Activator of protocatechuate pathway genes. LuxR cg2627 pcaO 3.64 26 family 3-oxoadipate enol-lactone hydrolase/4- cg2628 pcaC 3.97 27 carboxymuconolactonedecarboxylase 28 cg2629 pcaB 3-Carboxy-cis,cis-Muconate Cycloisomerase 3.74 29 cg2630 pcaG Protocatechuatedioxygenase alpha subunit 4.17 30 cg2631 pcaH Protocatechuatedioxygenase beta subunit 3.90 31 cg2633 - Putative restriction endonuclease 3.84 32 cg2634 catC Muconolactoneisomerase 4.57 33 cg2635 catB Chloromuconatecycloisomerase 3.75 34 cg2636 catA Catechol 1,2-dioxygenase 4.05 Benzoate 1,2-Dioxygenase Alpha Subunit (Aromatic cg2637 benA 4.21 35 ring hydroxylation dioxygenase A) 36 cg2638 benB Benzoate dioxygenase small subunit 3.87 ben cg2639 Benzoate dioxygenasereductase 4.55 37 C ben cg2640 Benzoate diol dehydrogenase bend 4.23 38 D 39 cg2641 benR Activator of catechol pathway genes. LuxR family 3.71 ben cg2642 Putative Benzoate Transport Protein 3.96 40 K 41 cg2643 benE Benzoate Membrane Transport Protein 3.48 a Gene ID according to the accession number BX927147 was used. b,c Gene name and annotation were obtained from the CoryneRegNet database (www.coryneregnet.de) and Brinkrolf et al., 2010 and Schröder and Tauch, 2010. d Expression ratios from a comparison of the H2O2-adapted strain under the oxidative stress condition vs. the wild-type strain under the no stress condition. Supplementary Table 5. List of the open reading frames (ORFs) showing a ≤ 2-fold decrease in comparison with those of the H2O2-adapted and wild-type strains.
Gene Expression No Gene nameb Annotationc IDa ratiod Peptidyl- 1 cg0048 ppiA prolylcis-trans 0.42 isomerase B putative 2 cg0092 - membrane 0.49 protein hypothetical cg0160 - 0.47 3 protein Methylated- DNA--protein- 4 cg0186 - cysteine 0.42 methyltransferas e hypothetical cg0282 csbD 0.25 5 protein Glycosyltransfer cg0394 - 0.43 6 ase hypothetical cg0440 - 0.19 7 protein 8 cg0470 - secreted protein 0.40 Phosphoglycero cg0482 gpmA 0.34 9 mutase 1 hypothetical cg0491 - 0.08 10 protein Extremely conserved cg0492 - 0.10 11 possible DNA- binding protein Hypothetical cg0493 - 0.12 12 protein Extremely conserved cg0494 - 0.32 13 hypothetical protein Activator of serine hydroxymethyltr cg0527 - 0.43 14 ansferase gene glyA.ArsR family cg0545 pitA putative low- 0.21 15 affinity phosphate transport protein Probable 50S ribosomal cg0573 rplL 16 subunit protein L7/L12 0.23 Elongation cg0583 fusA 17 factor G 0.47 Elongation cg0587 tuf 0.16 18 factor TU 30S ribosomal cg0599 rpsS 0.45 19 protein S19 Ribosomal cg0600 rplV 0.27 20 protein L22 30S ribosomal cg0601 rpsC 0.36 21 protein S3 50S ribosomal cg0602 rplP 0.10 22 protein L16 50S ribosomal cg0603 rpmC 0.46 23 protein L29 50S ribosomal cg0608 rplN 0.48 24 protein L14 Ribosomal cg0652 rpsM 0.35 25 protein S13 30S ribosomal cg0674 rpsI 0.31 26 protein S9 ABC-type transport 27 cg0737 - system, secreted 0.13 lipoprotein component O- 28 cg0755 metY Acetylhomoseri 0.35 ne (Thiol)-Lyase hypothetical cg0775 - 0.32 29 protein Acetyl/propiony l-CoA cg0812 dtsR1 0.32 30 carboxylase beta chain Ribosome- associated cg0867 - 0.29 31 protein Y (PSrp- 1) hypothetical cg0871 - 0.43 32 protein Activator of 33 cg0878 whcE thioredoxin 0.13 genes hypothetical cg0892 - 0.19 34 protein hypothetical cg0906 - 0.00 35 protein ABC-type cobalamin/Fe3+- siderophores 36 cg0924 - transport 0.36 system, periplasmic component DNA or RNA 37 cg0933 - helicase of 0.19 superfamily II 38 cg0958 - Secreted protein 0.23 3'- Phosphoadenosi ne 5'- cg0967 cysQ 0.46 39 phosphosulfate (PAPS) 3'- phosphatase 30S ribosomal cg0989 rpsN 0.44 40 protein S14 Anion-specific cg1109 porB 41 porin precursor 0.29 Enolase (2- 42 cg1111 eno phosphoglycerat e dehydratase) 0.44 Conserved protein/domain typically associated with cg1147 ssuI 43 flavoproteinoxy genases, DIM6/NTAB family 0.30 Thiol cg1236 tpx 44 peroxidase 0.43 ECF sigma factor involved 45 cg1271 sigE in responses to 0.09 cells surface stresses putative shikimate / cg1283 aroE1 0.38 46 quinate 5- dehydrogenase Homocysteinem cg1290 metE 0.22 47 ethyltransferase putative 48 cg1291 - membrane 0.41 protein Glyoxalase/Bleo mycin resistance/Dioxy cg1373 - 0.30 49 genase superfamily protein putative 50 cg1408 - membrane 0.35 protein 6- 51 cg1409 pfkA phosphofructoki 0.22 nase Putative 52 cg1479 glgP1 glycogen 0.28 phosphorylase Dephospho-coa cg1538 - 0.23 53 kinase hypothetical cg1556 - 0.24 54 protein putative cg1579 - 0.49 55 secreted protein 16S rRNA uridine-516 pseudouridylate 56 cg1615 - synthase or 0.24 related pseudouridylate synthase Putative signal transduction cg1630 - 0.06 57 protein, FHA domain Predicted 58 cg1633 - transcriptional 0.14 regulator Glyceraldehyde- 59 cg1791 gapA 3-phosphate 0.07 dehydrogenase Putative 60 cg1811 ihf integration host 0.34 factor CihF Putative cg1859 - 0.18 61 secreted protein hypothetical cg2027 - 0.28 62 protein cg2052 - putative 0.22 63 secreted protein hypothetical cg2079 - 0.31 64 protein Phosphocarrier cg2121 ptsH 0.37 65 protein HPR Ribosomal cg2167 rpsO 0.12 66 protein S15 putative secreted or cg2195 - 0.27 67 membrane protein 50S ribosomal cg2235 rplS 0.28 68 protein L19 Ankyrin repeat cg2254 - 0.21 69 protein Predicted metal- binding, 70 cg2274 - possibly nucleic 0.32 acid-binding protein hypothetical cg2283 - 0.46 71 protein 72 cg2291 pyk pyruvate kinase 0.45 putative cg2308 - 0.26 73 secreted protein Predicted 74 cg2320 - transcriptional 0.23 regulator hypothetical cg2325 - 0.47 75 protein putative 76 cg2362 - membrane 0.27 protein hypothetical cg2363 - 0.40 77 protein hypothetical protein cg2411 - 0.22 78 HesB/YadR/Yfh F family hypothetical cg2451 - 0.08 79 protein Predicted 80 cg2458 pgp2 phosphatase, 0.32 HAD family hypothetical cg2477 - 0.37 81 protein hypothetical cg2480 - 0.49 82 protein Bex protein (GTP-binding cg2510 bex 0.43 83 protein Era homolog) putative cg2518 - 0.36 84 secreted protein Secreted protein potentially 85 cg2561 thiX involved into 0.40 thiamin biosynthesis 30S ribosomal cg2573 rpsT 0.49 86 protein S20 Possible 87 cg2658 rpi phosphopentosei 0.05 somerase Putative dithiol- disulfide isomerase cg2661 - 0.49 88 involved in polyketide biosynthesis Membrane 89 cg2684 - protein DedA 0.29 family Bacterial regulatory cg2686 - 0.37 90 proteins, TetR family Cystathionine 91 cg2687 metB gamma- 0.45 Synthase putative 92 cg2701 - membrane 0.45 protein 93 cg2703 - Sugar permease 0.12 ABC-type sugar transport 94 cg2704 - system, 0.12 permease component Maltose-binding 95 cg2705 malE1 protein 0.30 precursor putative 96 cg2735 - membrane 0.37 protein O-acetylserine cg2833 cysK 0.04 97 (thiol)-lyase Predicted cg2835 - 0.41 98 acetyltransferase Butyryl- CoA:acetate cg2840 actA 0.32 99 coenzyme a transferase Conserved hypothetical cg2853 - 0.47 100 protein- fragment hypothetical cg2875 - 0.04 101 protein HIT family cg2880 - 0.27 102 hydrolase putative cg2952 - 0.33 103 secreted protein L-2,3- Butanediol cg2958 butA 0.22 104 dehydrogenase/a cetoinreductase 105 cg3008 porA Porin 0.27 hypothetical cg3009 - 0.03 106 protein Exodeoxyribonu cg3036 xthA 0.26 107 clease III Fructose- 108 cg3068 fba bisphosphateald 0.25 olase Beta- 109 cg3147 ‘bglY' glucosidase- 0.22 fragment ABC-type cobalamin/Fe3+- siderophores cg3148 - 0.24 110 transport system, ATPase component L-lactate cg3219 ldh 0.14 111 dehydrogenase hypothetical cg3257 - 0.14 112 protein putative 113 cg3260 - membrane 0.35 protein putative secreted cg3343 - 0.15 114 membrane protein putative 115 cg3409 thiD2 phosphomethylp 0.22 yrimidine kinase 50S ribosomal cg3432 rpmH 0.23 116 protein l34 hypothetical cg4000 - 0.08 117 protein hypothetical cg4007 - 0.00 118 protein a Gene ID according to the accession number BX927147 was used. b,c Gene name and annotation were obtained from the CoryneRegNet database (www.coryneregnet.de) and Brinkrolf et al., 2010 and Schröder and Tauch, 2010. d Expression ratios from the comparison of the H2O2-adapted strain under the oxidative stress condition vs. the wild-type strain under the no stress condition. Supplementary Table 6. Validation of selected gene expression profile via RT-qPCR at 4, 12, and 22 h of batch culture.
Gene Gene IDa Relative expressionc nameb 4 h 12 h 22 h Up-regulated WT HA HA WT HA HA WT HA HA in - - + - - + - - + RNA-seq d e f H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 1.00 2.00 1.40 1.00 3.20 4.80 1.00 ±.15 1.38 3.50 cg1748 - ±0.07 ±0.17 ±0.16 ±0.03 ±0.09 ±0.46 ±0.16 ±0.63 1.00 1.23 1.37 1.00 1.55 2.73 1.00 4.37 11.93 cg2610 - ±0.05 ±0.13 ±0.23 ±0.03 ±0.04 ±0.10 ±0.09 ±0.11 ±1.21 1.00 1.86 1.90 1.00 4.36 7.38 1.00 5.52 7.74 cg2618 vanK ±0.33 ±0.48 ±0.47 ±0.06 ±0.44 ±0.76 ±0.15 ±0.94 ±0.97 1.00 5.46 2.14 1.00 5.78 9.72 1.00 7.55 8.16 cg2622 pcaI ±0.20 ±0.52 ±0.18 ±0.14 ±0.73 ±1.01 ±0.18 ±0.95 ±1.10 1.00 1.27 1.38 1.00 1.87 3.06 1.00 2.01 1.28 cg2628 pcaC ±0.08 ±0.20 ±0.28 ±0.17 ±0.24 ±0.10 ±0.11 ±0.31 ±0.22 1.00 3.57 2.15 1.00 2.63 6.82 1.00 5.16 7.44 cg3420 sigM ±0.22 ±0.14 ±0.75 ±0.16 ±0.39 ±0.89 ±0.06 ±0.23 ±0.31 1.00 2.19 1.23 1.00 1.84 1.73 1.00 0.68 0.94 cg3422 trxB ±0.06 ±0.14 ±0.10 ±0.09 ±0.20 ±0.18 ±0.08 ±0.04 ±0.13 1.00 1.78 1.44 1.00 1.90 3.44 1.00 1.37 2.34 cg3423 trxC ±0.24 ±0.37 ±0.34 ±0.27 ±0.41 ±0.75 ±0.30 ±0.36 ±0.54 4h 12h 22h Down-regulated WT HA HA WT HA HA WT HA HA in - - + - - + - - + RNA-seq H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 H2O2 1.00 0.37 0.43 1.00 0.74 0.54 1.00 0.24 0.33 cg0878 whcE ±0.09 ±0.01 ±0.03 ±0.06 ±0.09 ±0.04 ±0.09 ±0.02 ±0.03 1.00 0.61 0.46 ±0.2 1.00 1.70 1.17 1.00 0.96 0.88 cg0933 - ±0.30 ±0.16 ±0.05 ±0.26 ±0.18 ±0.15 ±0.16 ±0.21 1.00 2.48 0.83 1.00 2.32 1.29 1.00 1.38 1.02 cg1271 sigE ±0.07 ±0.21 ±0.09 ±0.13 ±0.24 ±0.21 ±0.03 ±0.20 ±0.19 1.14 0.67 0.31 1.00 0.88 0.43 1.00 0.80 0.71 cg1791 gapA ±0.05 ±0.05 ±0.02 ±0.06 ±0.06 ±0.03 ±0.09 ±0.07 ±0.05 1.00 1.98 ±0.2 1.05 1.00 0.87 0.45 1.00 0.05 0.11 cg2833 cysK ±0.09 ±0.14 ±0.06 ±0.08 ±0.03 ±0.08 ±0.01 ±0.01 a Gene ID according to the accession number BX927147 was used. b Gene name was obtained from the CoryneRegNet database (www.coryneregnet.de) and references (Brinkrolf et al., 2010; Schröder and Tauch, 2010) c Relative expression resulted from cDNA synthesized using total RNA corresponding to 4, 12, and 22 h shown in Fig. 2. d, e, f WT and HA represent Corynebacterium glutamicum ATCC 13032 wild-type and the H2O2-adapted strain, respectively; - or + H2O2 indicate growth condition under the absence or presence of 10 mM H2O2, respectively.