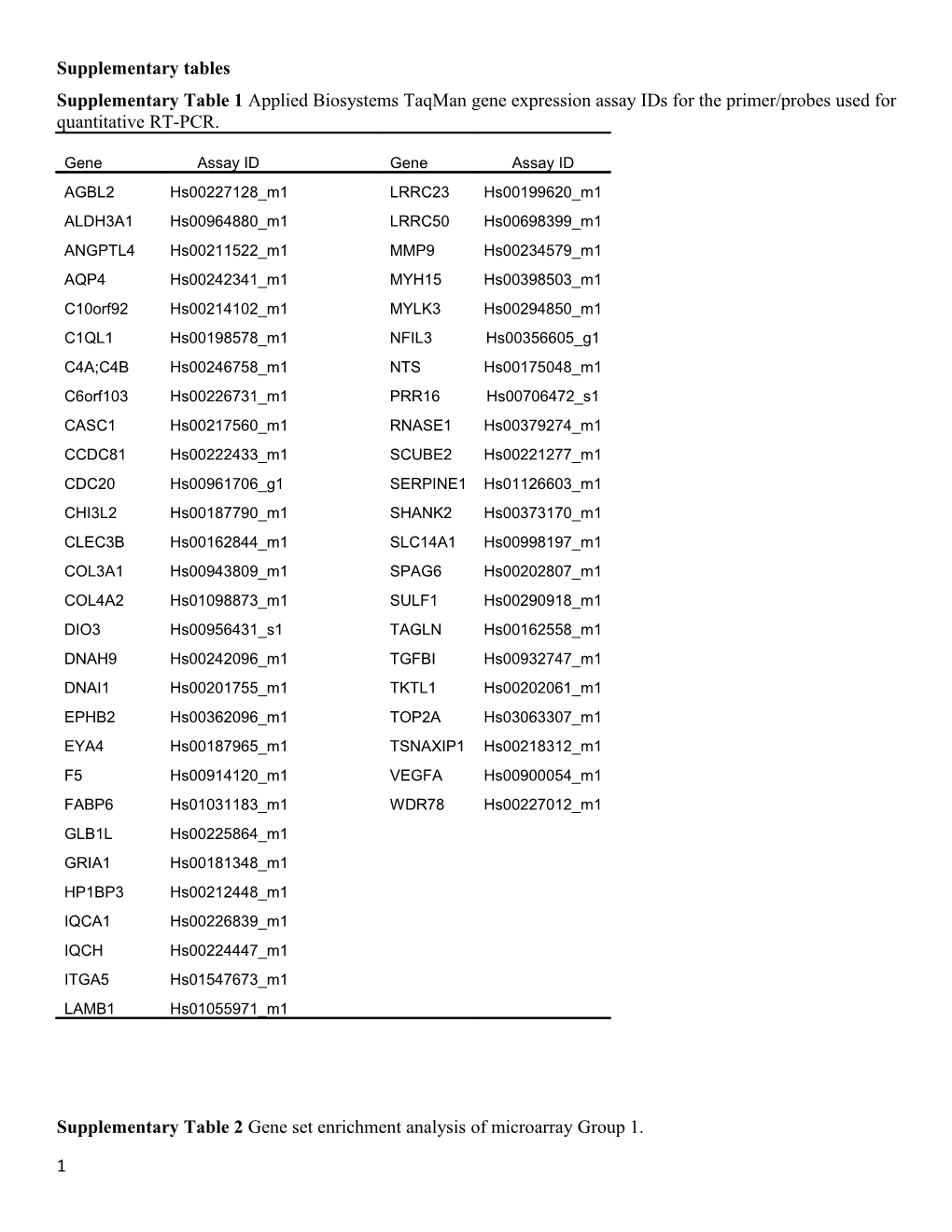
Supplementary tables Supplementary Table 1 Applied Biosystems TaqMan gene expression assay IDs for the primer/probes used for quantitative RT-PCR.
Gene Assay ID Gene Assay ID AGBL2 Hs00227128_m1 LRRC23 Hs00199620_m1 ALDH3A1 Hs00964880_m1 LRRC50 Hs00698399_m1 ANGPTL4 Hs00211522_m1 MMP9 Hs00234579_m1 AQP4 Hs00242341_m1 MYH15 Hs00398503_m1 C10orf92 Hs00214102_m1 MYLK3 Hs00294850_m1 C1QL1 Hs00198578_m1 NFIL3 Hs00356605_g1 C4A;C4B Hs00246758_m1 NTS Hs00175048_m1 C6orf103 Hs00226731_m1 PRR16 Hs00706472_s1 CASC1 Hs00217560_m1 RNASE1 Hs00379274_m1 CCDC81 Hs00222433_m1 SCUBE2 Hs00221277_m1 CDC20 Hs00961706_g1 SERPINE1 Hs01126603_m1 CHI3L2 Hs00187790_m1 SHANK2 Hs00373170_m1 CLEC3B Hs00162844_m1 SLC14A1 Hs00998197_m1 COL3A1 Hs00943809_m1 SPAG6 Hs00202807_m1 COL4A2 Hs01098873_m1 SULF1 Hs00290918_m1 DIO3 Hs00956431_s1 TAGLN Hs00162558_m1 DNAH9 Hs00242096_m1 TGFBI Hs00932747_m1 DNAI1 Hs00201755_m1 TKTL1 Hs00202061_m1 EPHB2 Hs00362096_m1 TOP2A Hs03063307_m1 EYA4 Hs00187965_m1 TSNAXIP1 Hs00218312_m1 F5 Hs00914120_m1 VEGFA Hs00900054_m1 FABP6 Hs01031183_m1 WDR78 Hs00227012_m1 GLB1L Hs00225864_m1 GRIA1 Hs00181348_m1 HP1BP3 Hs00212448_m1 IQCA1 Hs00226839_m1 IQCH Hs00224447_m1 ITGA5 Hs01547673_m1 LAMB1 Hs01055971_m1
Supplementary Table 2 Gene set enrichment analysis of microarray Group 1.
1 GENE SET NES FDR q-value p-value
ECM RECEPTOR INTERACTION 2.98 <0.001 <0.001
COMPLEMENT AND COAGULATION CASCADES 2.67 <0.001 <0.001
GLUTATHIONE METABOLISM 2.47 0.002 <0.001
PPAR SIGNALING PATHWAY 2.43 0.003 <0.001
TYPE I DIABETES MELLITUS 2.39 0.003 <0.001
FOCAL ADHESION 2.37 0.002 <0.001
CDMAC PATHWAY 2.32 0.003 <0.00
INTEGRIN PATHWAY 2.29 0.005 <0.001
CELL COMMUNICATION 2.28 0.004 <0.001
LAIR PATHWAY 2.25 0.005 0.001
NES: Normalized enrichment score. FDR: false discovery rate.
Supplementary Table 3 Expression of genes that define the mesenchymal phenotype in glioblastoma, in the microarray defined transcriptomal subgroups.
Fold change Group Gene 1 vs. Group 2 p-value
ANGPTL4 2.71 < 0.001
CHI3L1 8.09 < 0.001
COL4A1 4.31 < 0.001
COL4A2 4.08 < 0.001
MYL9 2.16 < 0.001
2 TIMP1 3.31 < 0.001
SERPINE1 2.62 < 0.001
FOSL2 2.27 < 0.001
PDLIM4 2.07 < 0.001
PDPN 1.36 0.028
TAGLN 3.84 < 0.001
PLA2G5 1.31 0.074
LIF 1.31 0.089
SFLJ25348 NA -
FAM20 NA -
NA, not available on U133 A microarray GeneChip
Supplementary Table 4 Gene set enrichment analysis of microarray Group 2.
GENE SET NES FDR q-value p-value
BASAL TRANSCRIPTION FACTORS -1.43 0.695 0.068
NAPHTHALENE AND ANTHRACENE DEGRADATION -1.35 0.518 0.124
AMINO PHOSPHONATE METABOLISM -1.23 0.571 0.185
RIBOFLAVIN METABOLISM -1.15 0.577 0.247
ATR BRCA PATHWAY -1.11 0.550 0.333
GLYCOSYL PHOSPHATIDYL INOSITOL ANCHOR BIOSYNTHESIS -0.98 0.704 0.481
DNA POLYMERASE -0.92 0.707 0.583
GAMMA HEXACHLORO CYCLOHEXANE DEGRADATION -0.88 0.687 0.585 3 GLYCOSPHINGOLIPID BIOSYNTHESIS NEOLACTO SERIES -0.81 0.724 0.740
NES: Normalized enrichment score. FDR: False discovery rate,
4 Supplementary Table 5 Clinical characteristics of patients used in the study.
Age at Extent of RFS Alive Dx Resectio WHO Adjuvan (weeks vs. OS LRS/ TOP2A ID (years) n Grade t XRT Recurrence ) Dead (weeks) SRS T/V IHC EP1 22 GT II No No 381 Alive 381 LRS T 0 EP2 28 GT II No No 396 Alive 396 LRS T 0 EP3 39 GT II No Yes 28 Alive 764 SRS T NA EP4 52 GT II No No 436 Alive 436 LRS T 0 EP5 2 GT III No Yes 66 Dead 68 SRS T NA EP6 6 GT II Yes No 424 Alive 424 LRS T 0 EP7 13 GT II Yes No 292 Alive 292 LRS T 0 EP8 44 GT II Yes No 260 Alive 260 LRS T 0 EP9 2 GT II Yes No 305 Alive 305 LRS T NA EP1 0 5 GT II Yes No 617 Alive 617 LRS T NA EP1 1 2 GT II No No 712 Alive 712 LRS V 0 EP1 2 2 GT II No No 696 Alive 696 LRS V 0 EP1 3 3 GT II No Yes 52 Dead 96 SRS V 1 EP1 4 12 GT II No Yes 76 Dead 184 SRS V 1 EP1 5 50 GT II No Yes 141 Alive 648 SRS V 0 EP1 6 51 GT II No No 254 Alive 254 LRS V 0 EP1 7 54 GT II No Yes 56 Alive 428 SRS V 0 EP1 8 15 GT III No Yes 16 Dead 116 SRS V 1 EP1 9 3 GT III No Yes 47 Dead 132 SRS V NA EP2 0 3 GT III No Yes 136 Dead 470 SRS V NA EP2 1 1 GT II Yes Yes 92 Alive 92 SRS V 0 EP2 2 2 GT II Yes No 216 Alive 216 LRS V 0 EP2 3 3 GT II Yes No 252 Alive 252 LRS V 1 EP2 4 5 GT II Yes No 240 Alive 240 LRS V 1 EP2 5 7 GT II Yes Yes 88 Alive 416 SRS V 0 EP2 6 10 GT II Yes No 244 Alive 244 LRS V 0 EP2 7 11 GT II Yes Yes 172 Alive 412 LRS V 0 EP2 8 11 GT II Yes No 248 Alive 248 LRS V 0 EP2 9 72 GT II Yes No 211 Alive 211 LRS V 0 EP3 0 1 ST II Yes Yes 104 Dead 288 SRS T 1 EP3 1 68 ST II Yes Yes 26 Dead 33 SRS T 0 5 EP3 2 10 ST II No No 284 Alive 284 LRS V 0 EP3 3 21 ST II No Yes 80 Alive 428 SRS V 1 EP3 4 29 ST II No Yes 92 Alive 184 SRS V NA EP3 5 5 ST II Yes Yes 120 Dead 160 SRS V 1 EP3 6 40 ST II Yes Yes 380 Alive 440 LRS V NA EP3 7 41 ST II Yes No 356 Alive 586 LRS V NA
6 Age at Alive Dx Extent of WHO Adjuvant RFS vs. OS LRS/ TOP2A ID (years) Resection Grade XRT Recurrence (weeks) Dead (weeks) SRS T/V IHC EP38 58 ST II Yes No 264 Alive 264 LRS V 0 EP39 67 ST II Yes No 416 Alive 416 LRS V NA EP40 3 ST II No Yes 28 Dead 332 SRS V 1 EP41 10 GT III No Yes 64 Dead 113 SRS T NA EP42 10 GT III No Yes 43 Dead 112 SRS T NA EP43 3 GT II No Yes 129 Dead 228 SRS V 1 EP44 11 GT III No Yes 139 Dead 198 SRS V NA EP45 4 GT II Yes Yes 26 Alive 169 SRS V 1 EP46 0 ST II No Yes 48 Dead 328 SRS V 1 EP47 1 ST II No Yes 175 Dead 306 LRS V 0 EP48 1 ST III No Yes 56 Dead 64 SRS V 0 EP49 7 ST III Yes Yes 28 Alive 192 SRS V 1 EP50 2 Unknown II No Yes 225 Dead 456 LRS V 1 EP51 4 Unknown II No Yes 27 Alive 1054 SRS V 1 Unknow EP52 1 GT II n Yes 104 Alive 207 SRS T NA Unknow EP53 5 GT III n Yes 25 Dead 162 SRS T NA Unknow EP54 8 GT II n No 475 Alive 475 LRS T NA EP55 0 GT II No Yes 88 Dead 184 SRS V 1 Unknow EP56 4 ST II n Yes 136 Alive 271 SRS T NA
Dx: diagnosis, GT: Gross total resection, ST: Subtotal resection, RFS: recurrence-free survival, defined as the time between initial surgery and recurrence, OS: overall survival, defined as the time between surgery and death or the last available follow up, T: testing dataset, V: validation dataset, TOP2A IHC: 0 = low expression 1= high expression NA= data not available.
7 Supplementary Table 6 The survival associated 51-gene expression signature of infratentorial ependymoma.
Genes over expressed in SRS Genes over expressed in LRS Fold change Fold change Fold change microarray Group 1 Fold change microarray Group 1 Gene SRS vs. LRS vs. Group 2 Gene SRS vs. LRS vs. Group 2 PRR16 17.96 2.15 NTS 0.03 0.22 MMP9 16.86 2.32 SLC14A1 0.04 0.64 COL3A1 14.65 4.05 AGBL2 0.09 0.28 RNASE1 11.8 0.96 ALDH3A1 0.10 0.45 LAMB1 10.74 2.35 FABP6 0.13 0.41 DIO3 8.54 1.64 MYLK3 0.14 0.50 COL4A2 6.90 4.14 LRRC23 0.14 0.46 EPHB2 5.90 1.13 CHI3L2 0.15 4.77 CDC20 5.73 1.55 C10orf92 0.16 0.28 TOP2A 5.57 2.89 TSNAXIP1 0.17 0.49 SERPINE1 5.45 3.06 C6orf103 0.17 0.25 TKTL1 5.38 3.17 F5 0.20 0.94 SCUBE2 4.92 1.42 LRRC50 0.23 0.30 HP1BP3 3.79 1.03 SHANK2 0.24 0.20 C1QL1 3.78 1.99 IQCH 0.25 0.42 ANGPTL4 3.49 2.71 IQCA1 0.28 0.43 NFIL3 3.37 2.21 CLEC3B 0.28 0.43 VEGFA 3.19 5.52 SULF1 0.31 0.88 TAGLN 3.15 3.84 WDR78 0.31 0.49 TGFBI 3.04 4.11 MYH15 0.31 0.51 C4A;C4B 2.93 2.54 EYA4 0.31 0.46 ITGA5 2.50 2.60 GLB1L 0.33 0.61 CCDC81 0.33 0.43 DNAH9 0.34 0.39 AQP4 0.34 1.31 DNAI1 0.37 0.36 GRIA1 0.37 0.48 SPAG6 0.40 0.36 CASC1 0.40 0.37
SRS: short recurrence-free survivors, LRS: long recurrence-free survivors Supplementary Table 7 Comparison of transcriptomal subgroups with survival associated subgroups.
Group 1 Group 2 p-value*
8 LRS-associated genes 2 27 < 0.001 SRS-associated genes 20 0
* Fisher’s exact test two-tailed. SRS: short recurrence-free survivors, LRS: long recurrence-free survivors
Supplementary Table 8 Univariate Cox regression analysis of the transcriptomal subgroups as defined by the genes from Fig. 1b as applied to the Heidelberg dataset from Witt et al.
Recurrence-free survival Overall survival
Variable HR p-value HR p-value Transcriptomal subgroup 0.33 0.041 0.07 0.015
9