Chapter 19 – Gene Manipulation
Outline:
Mutagenesis In-vitro mutagenesis Cassette mutagenesis PCR site-directed mutagenesis Gene Knockout Gene Silencing
Overview:
FIGURE 1: MUTAGENESIS
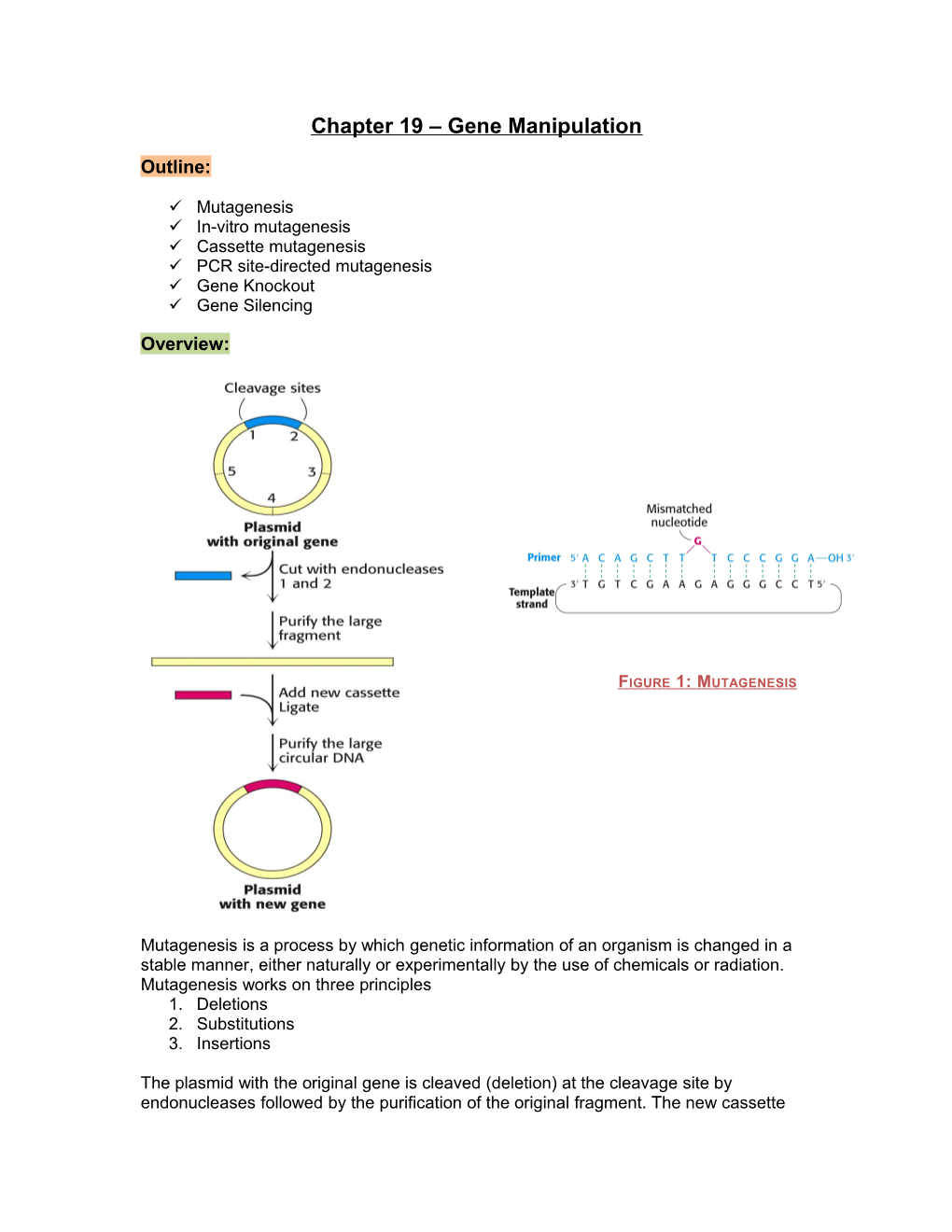
Mutagenesis is a process by which genetic information of an organism is changed in a stable manner, either naturally or experimentally by the use of chemicals or radiation. Mutagenesis works on three principles 1. Deletions 2. Substitutions 3. Insertions
The plasmid with the original gene is cleaved (deletion) at the cleavage site by endonucleases followed by the purification of the original fragment. The new cassette ligate will then by inserted followed by purification of circular DNA, which leads to the production of plasmid with the new gene.
FIGURE 2: GENERAL STRATEGY OF IN-VITRO MUTAGENESIS
In-vitro mutagenesis is the production of either random or specific mutations in a piece of cloned DNA. Then the DNA is introduced in a cell to assess the results.
(i) Plasmid DNA is mutagenized in vitro, then introduced into E. coli by transformation. Mutant clones can be isolated and tested individually or a library of mutant plasmids can be tested using genetic screening.
(ii) Creating a mutation by manipulation of a restriction site: Plasmid DNA cut with EcoRI. Treatment with S1 and ligase produces a 4bp deletion mutant. Treatment with DNA polymerase and ligase produces a 4bp insertion mutant. EcoR1 recognizes GAATTC FIGURE 3: CASSETTE MUTAGENESIS
Cassette mutagenesis is a process in which a wild type DNA fragment is being replaced by a DNA fragment of desired mutation.
(i) Plasmid DNA (a wild type sequence) is cut by two restriction enzymes HindIII and EcoR1. A DNA fragment (cassette) containing the desired mutation is introduced through DNA ligase. The mutant DNA formed consists of the wild type DNA and the new mutated fragment, which is then transformed into E.Coli for a specific mutation.
(ii) Cassette mutagenesis using doped oligonucleotides (each bottle is contaminated with small amount of other three precursors i.e. AGCT) to generate numerous mutants in a single experiment. A 30 base pair sequence with mutation at each position can be obtained. It is a very powerful technique which produces thousands of mutants (one or multiple) sequencing each one of them. FIGURE 4: PCR BASED SITE-DIRECTED MUTAGENESIS. TAC (FOR TYROSINE) WILL PRODUCE TTC (FOR PHENYLALANINE)
PCR (Polymerase Chain Reaction) is a site directed mutagenesis using double stranded DNA.
As shown in the figure above, the parental plasmid consists of double stranded E.Coli plasmid. The two DNA strands are separated by denaturing the parental plasmid and mutagenic primers are added to produce new mutants.
Polymerase chain reaction will then produce numerous daughter plasmids without methyl groups and a numerous parental plasmid (wild type) without mutant. Treatment with restriction enzyme Dpnl will cut the parental plasmid at the methylated position which will not transform into any mutation.
The daughter plasmid will be the mutated plasmid which will be used to transform into E.Coli to produce desired mutation. FIGURE 5: GENE REPLACEMENT IN YEAST
1) A deletion or mutant gene his3 is inserted into a yeast vector HIS3.
2) Diploid yeast is transformed with exogenous DNA. Recombinant plasmid integrates with XV yeast chromosome and is fairly an automated process.
3) Homologous recombination occurs and a heterozygous yeast cell is generated
4) Intrachromosomal recombination and excision of URA3 will form original gene or produce a new mutant gene. The wild type gene can be replaced or remains the same. FIGURE 6: GENE KNOCKOUT IN YEAST
Gene knockout is a genetic technique in which an organism is engineered to carry genes that are made inoperative. The method works in the following ways:
1) A deletion or mutant gene is inserted into a yeast vector (wild type).
2) Diploid yeast is transformed with exogenous DNA.
3) Homologous recombination occurs and a heterozygous yeast cell is generated.
4) The heterozygous yeast undergoes meiosis (Sporulation is the change from diploid to haploid).
5) The 4 haploid spores are tested for viability and other phenotypes.
Out of the four haploid spores, two are viable and the other two are inviable. The viable spores are the wild type ones which will be able to survive and the inviable ones will be the knock out ones which will not be able to survive after disruption. Gene knockout in mice
ES (embryonic stem cells) heterozygous for X, homozygous for marker (black coat) transplant into blastocoel cavity of 4.5 days embryos (homozygous for white coat). Early embryo implant into a pseudopregnant female.
Transgenic mice
Cell-type-specific gene knockouts in mice To determine the function of these genes, it is possible to replace an organism’s wild type gene with an inactive gene to create a “gene knockout” It is also possible to introduce additional genes (transgenes) to create a transgenic organism
FIGURE 7: PROCEDURE FOR PRODUCING GENE-TARGETED KNOCKOUT MOUSE
FIGURE 8: TRANSGENIC PLANTS (1) Transgenic plants are plants possessing a single or multiple genes transferred from a different species. It refers to plants created in lab using recombinant DNA technology. Tumor inducing plasmids can be used to introduce new genes into plant cells. The T- DNA segment of Ti plasmids can be integrated into plant genome.
FIGURE 9: FORMATION OF A CROWN GALL
Crown gall tumors: Formation of a crown gall tumor in plant. Agro bacterium cells enter a wound in the plant, usually at the crown, or the junction of the root and stem. Bacteria have Ti plasmid containing a segment (red) that promotes tumor formation in infected plants, the Ti gene direct the formation of a crown gall, which nourishes the invading bacteria. FIGURE 10: RNAI (RNA INTERFERENCE) MODEL
In this model, the proteins Dicer is shown "dicing" a long double-stranded (double stranded RNA) into small pieces called short-interfering RNAs (siRNAs). One strand of these siRNAs is used by the RNAi Silencing Complex to target and destroy a specific gene (mRNA). The efforts here are focused on studying how RNAi works in mammals such as mice and humans. Exactly how mammalian RNAi works is still a mystery. Many proteins are probably involved, and knowing their identities will help unravel the mystery of RNAi.
FIGURE 11: RNAI SUPPRESSES GENES
The short siRNA pieces unwind into single strand RNAs, which then combine with proteins to form a complex called RISC (RNA-Induced Silencing Complex). The RISC then captures a native mRNA molecule that complements the short siRNA sequence. If the pairing (native mRNA and siRNA piece) is essentially perfect, the native mRNA is cut into useless RNA fragments that aren't translated. If however, the pairing is less than perfect then the RISC complex binds to the mRNA and blocks ribosome movement along the native mRNA also halting translation. The net effect is NO PROTEIN IS MADE. FIGURE 12: EXAMPLE OF RNAI (NATURE 391: 806-11)
The above figure shows the effects of mex-3 RNA interference on levels of the endogenous mRNA. Nomarski DIC micrographs show in situ hybridization of 4-cell stage embryos.
(A) Negative control showing lack of staining in the absence of the hybridization probe. (B) Embryo from uninjected parent showing normal pattern of endogenous mex-3 RNA (purple staining). (C) Embryo from parent injected with purified mex-3 antisense RNA. These embryos (and the parent animals) retain mex-3 mRNA, although levels may be somewhat less than wild type. (D) Late 4-cell stage embryo from a parent injected with dsRNA corresponding to mex-3; no mex-3 RNA is detected.
Gene Silencing
• Gene silencing is a general term describing epigenetic processes of gene regulation
• The term gene silencing is generally used to describe the "switching off" of a gene by a mechanism other than genetic modification. That is, a gene which would be expressed (turned on) under normal circumstances is switched off by machinery in the cell
In two papers published in the online edition of the journal Nature Chemical Biology, researchers at UT Southwestern Medical Center say they have developed a technique that can control gene expression by turning them on or off at the DNA level. In doing so, the research team may have paved the way for the development of new drugs designed to treat many serious diseases. As diagnoses and gene research improves, scientists are consistently finding that nearly all diseases start at the genetic level. "Virtually every disease starts at the level of malfunctioning gene expression, or viral or bacterial gene expression," said Dr. David Corey, professor of pharmacology and biochemistry (2). References:
1. http://www.anselm.edu/homepage/jpitocch/genbio/plantplasmidtechn.JPG
2. http://www.scienceagogo.com/news/20050702030521data_trunc_sys.shtml