For publication Supplement material
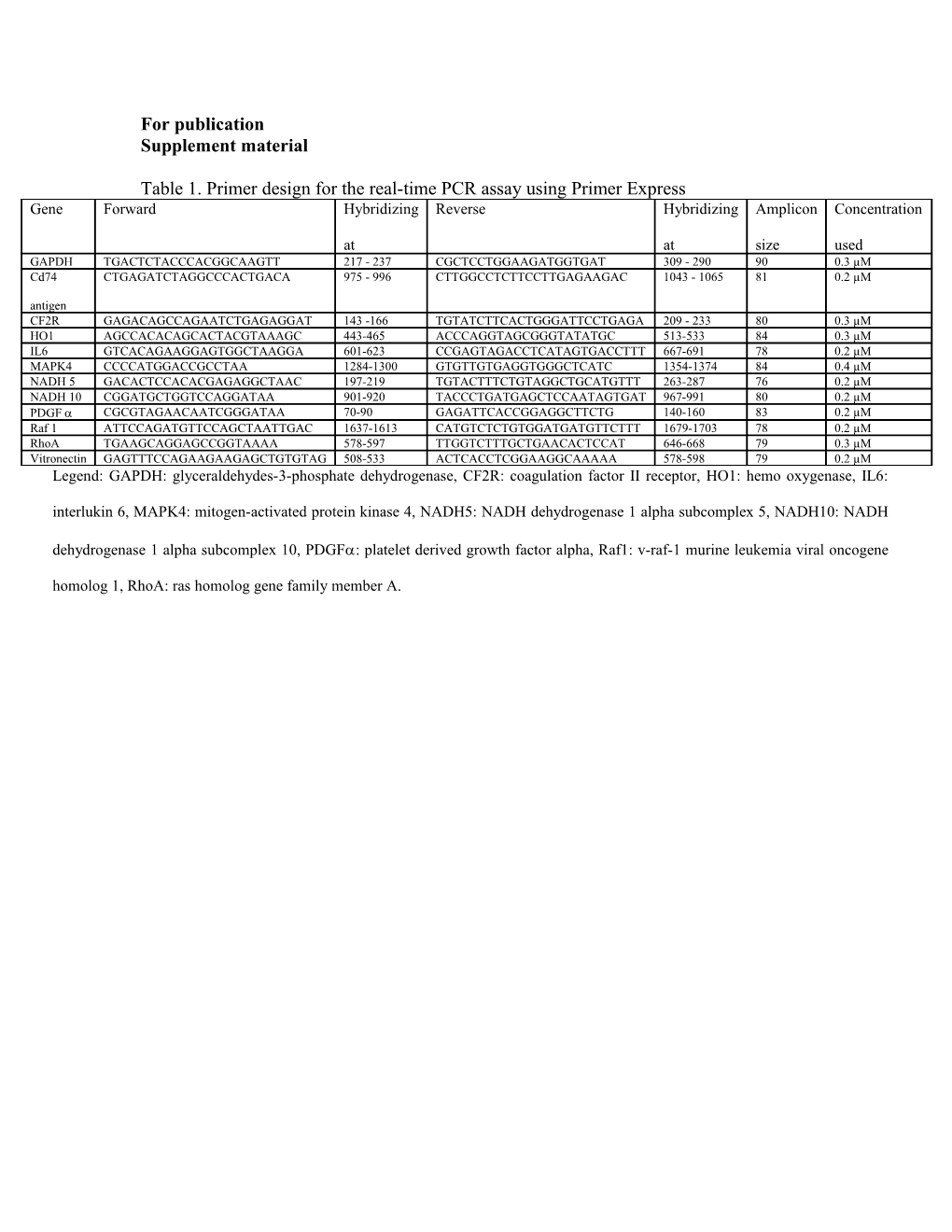
Table 1. Primer design for the real-time PCR assay using Primer Express Gene Forward Hybridizing Reverse Hybridizing Amplicon Concentration
at at size used GAPDH TGACTCTACCCACGGCAAGTT 217 - 237 CGCTCCTGGAAGATGGTGAT 309 - 290 90 0.3 µM Cd74 CTGAGATCTAGGCCCACTGACA 975 - 996 CTTGGCCTCTTCCTTGAGAAGAC 1043 - 1065 81 0.2 µM antigen CF2R GAGACAGCCAGAATCTGAGAGGAT 143 -166 TGTATCTTCACTGGGATTCCTGAGA 209 - 233 80 0.3 µM HO1 AGCCACACAGCACTACGTAAAGC 443-465 ACCCAGGTAGCGGGTATATGC 513-533 84 0.3 µM IL6 GTCACAGAAGGAGTGGCTAAGGA 601-623 CCGAGTAGACCTCATAGTGACCTTT 667-691 78 0.2 µM MAPK4 CCCCATGGACCGCCTAA 1284-1300 GTGTTGTGAGGTGGGCTCATC 1354-1374 84 0.4 µM NADH 5 GACACTCCACACGAGAGGCTAAC 197-219 TGTACTTTCTGTAGGCTGCATGTTT 263-287 76 0.2 µM NADH 10 CGGATGCTGGTCCAGGATAA 901-920 TACCCTGATGAGCTCCAATAGTGAT 967-991 80 0.2 µM PDGF CGCGTAGAACAATCGGGATAA 70-90 GAGATTCACCGGAGGCTTCTG 140-160 83 0.2 µM Raf 1 ATTCCAGATGTTCCAGCTAATTGAC 1637-1613 CATGTCTCTGTGGATGATGTTCTTT 1679-1703 78 0.2 µM RhoA TGAAGCAGGAGCCGGTAAAA 578-597 TTGGTCTTTGCTGAACACTCCAT 646-668 79 0.3 µM Vitronectin GAGTTTCCAGAAGAAGAGCTGTGTAG 508-533 ACTCACCTCGGAAGGCAAAAA 578-598 79 0.2 µM Legend: GAPDH: glyceraldehydes-3-phosphate dehydrogenase, CF2R: coagulation factor II receptor, HO1: hemo oxygenase, IL6:
interlukin 6, MAPK4: mitogen-activated protein kinase 4, NADH5: NADH dehydrogenase 1 alpha subcomplex 5, NADH10: NADH
dehydrogenase 1 alpha subcomplex 10, PDGF: platelet derived growth factor alpha, Raf1: v-raf-1 murine leukemia viral oncogene
homolog 1, RhoA: ras homolog gene family member A. Table 2. DAVID analysis for CAEC – The main effect of antioxidant (37 genes were analysed) Annotation cluster 1 (N=6) Enrichment score: 2.16 Gene Name Genbank_Accession Breast cancer anti-estrogen resistance 1 NM_012931 Fas (tnf receptor superfamily, member 6) D26113 Nucleophosmin M37041 Phosphofructokinase, NM_013190 Protein kinase, camp dependent regulatory, type i, alpha M17086 V-raf-leukemia viral oncogene 1 NM_012639 Term Count (n) Pvalue Kinase binding 5 <0.01 Enzyme binding 6 0.01 Annotation cluster 2 (N=13) Enrichment score: 1.49 Adenylate cyclase 4 NM_019285 Breast cancer anti-estrogen resistance 1 NM_012931 Chemokine receptor 5 Y12009 Cytochrome c oxidase subunit iv isoform 1 J05425, NM_017202 Early growth response 1 J04154 Endothelin receptor type b S65355 Fas (tnf receptor superfamily, member 6) D26113 Lecithin cholesterol acyltransferase NM_017024 Phosphofructokinase, liver NM_013190 Proteasome subunit, beta type 2 D21799 Protein kinase, camp dependent regulatory, type i, alpha M17086 V-raf-leukemia viral oncogene 1 NM_012639 Xanthine dehydrogenase NM_017154 Term Count (n) Pvalue Response to organic substance 9 <0.01 Response to endogenous stimulus 6 0.02 Response to chemical stimulus 13 0.04 Response to hormone stimulus 5 0.05 Annotation cluster 3 (N=7) Enrichment score: 1.37 Adenylate cyclase 4 NM_019285 Endothelin receptor type b S65355 Nucleophosmin M37041 Proteasome activator subunit 1 D45249 Proteasome subunit, alpha type 2 NM_017279 Proteasome subunit, beta type 2 D21799 Protein kinase, camp dependent regulatory, type i, alpha M17086 Term Count (n) Pvalue Positive regulation of catalytic activity 7 <0.01 Positive regulation of molecular function 7 <0.01 Regulation of catalytic activity 7 0.01 Regulation of molecular function 7 0.02 Annotation cluster 4 (N=24) Enrichment score: 1.32 Adenylate cyclase 4 NM_019285 Breast cancer anti-estrogen resistance 1 NM_012931 Calbindin 1 M31178 Chemokine receptor 5 Y12009 J05425, Cytochrome c oxidase subunit iv isoform 1 NM_017202 Dynein light chain roadblock-type 1 AY026512 Early growth response 1 J04154 Endothelin receptor type b S65355 Fas (tnf receptor superfamily, member 6) D26113 Glutamate decarboxylase 1 NM_017007 Lecithin cholesterol acyltransferase NM_017024 Ninjurin 2 AB040815 Nucleophosmin M37041 Phosphofructokinase NM_013190 Prosaposin M19936 Proteasome activator subunit 1 D45249 Proteasome subunit, alpha type 2 NM_017279 Proteasome subunit, beta type 2 D21799 Protein kinase, camp dependent regulatory, type i, alpha M17086 Solute carrier family 2 member 4 NM_012751 Syntaxin binding protein 1 NM_013038 Transcription factor 12 NM_013176 V-raf-leukemia viral oncogene 1 NM_012639 Xanthine dehydrogenase NM_017154 Term Count (n) Pvalue Response to stimulus 18 0.01 Response to chemical stimulus 13 0.04 Biological regulation 21 0.04 Annotation cluster 5 (N=23) Enrichment score: 1.31 Adenylate cyclase 4 NM_019285 Breast cancer anti-estrogen resistance 1 NM_012931 Calbindin 1 M31178 Carboxypeptidase d NM_012836 Chemokine receptor 5 Y12009 Early growth response 1 J04154 Endothelin receptor type b S65355 Fas (tnf receptor superfamily, member 6) D26113 Inter-alpha trypsin inhibitor, heavy chain 3 NM_017351 Lecithin cholesterol acyltransferase NM_017024 Lysosomal-associated membrane protein 1 M34959 Nucleophosmin M37041 Phosphofructokinase NM_013190 Prosaposin M19936 Proteasome activator subunit 1 D45249 Proteasome subunit, alpha type 2 NM_017279 Proteasome subunit, beta type 2 D21799 Protein kinase, camp dependent regulatory, type i, alpha M17086 Similar to 40s ribosomal protein s15 NM_017151 Syntaxin binding protein 1 NM_013038 Transcription factor 12 NM_013176 V-raf-leukemia viral oncogene 1 NM_012639 Xanthine dehydrogenase NM_017154 Term Count (n) Pvalue Positive regulation of catalytic activity 7 <0.01 Positive regulation of molecular function 7 <0.01 Regulation of protein modification process 5 0.01 Regulation of catalytic activity 7 0.01 Negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.01 Anaphase-promoting complex-dependent proteasomal ubiquitin- dependent protein catabolic process 3 0.01 Negative regulation of ubiquitin-protein ligase activity 3 0.01 Negative regulation of ligase activity 3 0.01 Psitive regulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.01 Reulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.01 Positive regulation of ubiquitin-protein ligase activity 3 0.01 Positive regulation of ligase activity 3 0.01 Positive regulation of protein modification process 4 0.01 Negative regulation of protein ubiquitination 3 0.01 Negative regulation of cellular process 10 0.01 Regulation of ubiquitin-protein ligase activity 3 0.01 Regulation of ligase activity 3 0.01 Cell cycle process 5 0.01 Positive regulation of protein ubiquitination 3 0.02 Regulation of molecular function 7 0.02 Proteasomal protein catabolic process 3 0.02 Proteasomal ubiquitin-dependent protein catabolic process 3 0.02 Positive regulation of cellular protein metabolic process 4 0.02 Negative regulation of biological process 10 0.02 Regulation of protein ubiquitination 3 0.02 Positive regulation of protein metabolic process 4 0.02 Regulation of cellular protein metabolic process 5 0.02 Negative regulation of catalytic activity 4 0.02 Negative regulation of protein modification process 3 0.03 Cell cycle 5 0.03 Regulation of protein metabolic process 5 0.04 Negative regulation of molecular function 4 0.04 Annotation cluster 6 (N=13) Enrichment score: 1.11 Calbindin 1 M31178 Chemokine receptor 5 Y12009 Chloride channel 1 NM_013147 Dynein light chain roadblock-type 1 AY026512 Early growth response 1 J04154 Endothelin receptor type b S65355 Fas (tnf receptor superfamily, member 6) D26113 Glutamate decarboxylase 1 NM_017007 Nucleophosmin M37041 Phosphofructokinase NM_013190 Prosaposin M19936 Syntaxin binding protein 1 NM_013038 Xanthine dehydrogenase NM_017154 Term Count (n) Pvalue Regulation of neurological system process 4 0.01 Regulation of multicellular organismal process 7 0.05 Annotation cluster 7 (N=14) Enrichment score: 1.02 Acetyl-coenzyme a acetyltransferase 1 D00512 Adenylate cyclase 4 NM_019285 Calbindin 1 M31178 Calsequestrin 2 NM_017131 Carboxypeptidase d NM_012836 Chloride channel 1 NM_013147 Early growth response 1 J04154 Farnesyl diphosphate farnesyl transferase 1 NM_019238 Glutamate decarboxylase 1 NM_017007 Nucleobindin 2 AF250142 Phosphofructokinase NM_013190 Potassium inwardly-rectifying channel, subfamily j, member 3 U09243 V-raf-leukemia viral oncogene 1 NM_012639 Xanthine dehydrogenase NM_017154 Term Count (n) Pvalue Ion binding 13 0.04
N, total number of genes in the cluster group. n, total number of genes under the GO term Table 3. DAVID analysis for CAEC – The main effect of Ex (35 genes were analysed)
Annotation cluster 1 (N=23) Enrichment score: 1.65 Adenylate cyclase 4 NM_019285 Adrenergic, beta, receptor kinase 1 M87854 Cathepsin h M38135 Enolase 3, beta, muscle Y00979 Fucosyltransferase 2 AB006138 Glutathione s-transferase, theta 2 NM_012796 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Inositol 1,4,5-trisphosphate 3-kinase a X56917 Insulin degrading enzyme NM_013159 Inter-alpha trypsin inhibitor, heavy chain 3 NM_017351 Lactalbumin, alpha NM_012594 Mitogen-activated protein kinase 4 Z21935 Mitogen-activated protein kinase 8 interacting protein 1 AF109774 N-acylaminoacyl-peptide hydrolase NM_012500 Nuclear factor i/b AB012231 Proteasome subunit, alpha type 5 D10756 Protein interacting with prkca 1 AJ240083 Protein kinase, amp-activated, gamma 1 non-catalytic subunit NM_013010 Rab geranylgeranyltransferase, beta subunit S62097 Similar to 40s ribosomal protein s26 NM_013224 Similar to elongation factor 2 Y07504 Similar to ribosomal protein l6 S71021 Smad family member 2 Nm_019191 Term Count (n) Pvalue Primary metabolic process 23 <0.01 Protein metabolic process 14 0.01 Cellular protein metabolic process 12 0.01 Metabolic process 23 0.02 Cellular metabolic process 20 0.02 Macromolecule metabolic process 17 0.03 Annotation cluster 2 (N=8) Enrichment score: 1.51 Adenylate cyclase 4 NM_019285 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Mitogen-activated protein kinase 8 interacting protein 1 AF109774 Proteasome subunit, alpha type 5 D10756 Protein interacting with prkca 1 AJ240083 Protein kinase, amp-activated, gamma 1 non-catalytic subunit NM_013010 Ras homolog gene family, member a; ras homolog gene family, member c AY026069 Smad family member 2 NM_019191 Term Count (n) Pvalue Regulation of catalytic activity 7 0.01 Enzyme binding 5 0.03 Protein kinase binding 3 0.05 Annotation cluster 3 (N=16) Enrichment score: 1.36 Adenylate cyclase 4 NM_019285 Adrenergic, beta, receptor kinase 1 M87854 Atp-binding cassette, sub-family b, member 4 NM_012690 Fucosyltransferase 2 AB006138 Glutathione s-transferase, theta 2 NM_012796 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Inositol 1,4,5-trisphosphate 3-kinase a X56917 Insulin degrading enzyme NM_013159 Lactalbumin, alpha NM_012594 Mitogen-activated protein kinase 4 Z21935 Protein interacting with prkca 1 AJ240083 Protein kinase, amp-activated, gamma 1 non-catalytic subunit NM_013010 Rab geranylgeranyltransferase, beta subunit S62097 Ras homolog gene family, member a; ras homolog gene family, member c AY026069 Similar to elongation factor 2 Y07504 Smad family member 2 NM_019191 Term Count (n) Pvalue Purine ribonucleotide binding 10 0.01 Ribonucleotide binding 10 0.01 Purine nucleotide binding 10 0.01 Transferase activity 9 0.02 Adenyl ribonucleotide binding 8 0.03 Nucleotide binding 10 0.03 Adenyl nucleotide binding 8 0.04 Purine nucleoside binding 8 0.04 Nucleoside binding 8 0.04 Annotation cluster 4 (N=8) Enrichment score: 1.19 Atp-binding cassette, sub-family b, member 4 NM_012690 Cathepsin h M38135 Insulin degrading enzyme NM_013159 N-acylaminoacyl-peptide hydrolase NM_012500 Proteasome subunit, alpha type 5 D10756 Protein interacting with prkca 1 AJ240083 Ras homolog gene family, member a; ras homolog gene family, member c AY026069 Similar to elongation factor 2 Y07504 Term Count Pvalue Nucleoside-triphosphatase activity 5 0.04 Pyrophosphatase activity 5 0.04 Hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides 5 0.04 Hydrolase activity, acting on acid anhydrides 5 0.05
N, total number of genes in the cluster group. n, total number of genes under the GO term Table 4. DAVID analysis for CAEC - The interaction effect (38 genes were analysed)
Annotation cluster 1 (N=17) Enrichment score: 3.12 Genbank Gene name accesion 3-hydroxy-3-methylglutaryl-coenzyme a synthase 1 X52625 Aminolevulinate, delta-, synthase 2 NM_013197 B-cell translocation gene 3 NM_019290 Collagen, type i, alpha 1 M12199 Gamma-glutamyltransferase 5 NM_019235 Glutamate-ammonia ligase M29599 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Inhibitor of dna binding 2 NM_013060 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Polymerase, beta NM_017141 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Solute carrier family 6, member 3 M80233 Timp metallopeptidase inhibitor 2 AJ409332 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Response to organic substance 13 <0.01 Response to endogenous stimulus 8 <0.01 Response to chemical stimulus 16 <0.01 Response to stimulus 17 0.04 Annotation cluster 2 (N=25) Enrichment score: 2.54 Aminolevulinate, delta-, synthase 2 NM_013197 B-cell translocation gene 3 NM_019290 Collagen, type i, alpha 1 M12199 Glutamate decarboxylase 1 NM_017007 Glutamate-ammonia ligase M29599 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase (decycling) 1 NM_012580 Homer homolog 1 AB007688 Inhibitor of dna binding 2 NM_013060 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Lectin, galactoside-binding, soluble, 9 U59462 Mal, t-cell differentiation protein NM_012798 Peripheral myelin protein 22 NM_017037 Polymerase, beta NM_017141 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Rab9a, member ras oncogene family AF325692 Signal-regulatory protein alpha D85183 Solute carrier family 6, member 3 M80233 Sparc-like 1 NM_012946 Timp metallopeptidase inhibitor 2 AJ409332 Transcription factor 12 NM_013176 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Negative regulation of biological process 13 <0.01 Negative regulation of cellular process 12 <0.01 Biological regulation 25 <0.01 Regulation of cell proliferation 8 <0.01 Regulation of biological process 22 0.01 Regulation of cellular process 21 0.01 Annotation cluster 3 (N=15) Enrichment score: 2.37 B-cell translocation gene 3 NM_019290 Glutamate-ammonia ligase M29599 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Inhibitor of dna binding 2 NM_013060 Lectin, galactoside-binding, soluble, 9 U59462 Mal, t-cell differentiation protein NM_012798 Peripheral myelin protein 22 NM_017037 Prostaglandin-endoperoxide synthase 2 NM_017232 Signal-regulatory protein alpha D85183 Solute carrier family 6, member 3 M80233 Timp metallopeptidase inhibitor 2 AJ409332 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Regulation of multicellular organismal process 10 <0.01 Regulation of cell proliferation 8 <0.01 Positive regulation of cellular process 10 0.04 Annotation cluster 4 (N=5) Enrichment score: 2.26 Aminolevulinate, delta-, synthase 2 NM_013197 Gamma-glutamyltransferase 5 NM_019235 Glutathione s-transferase, theta 2 NM_012796 Heme oxygenase 1 NM_012580 Hydroxymethylbilane synthase X06827 Term Count Pvalue Heme metabolic process 3 <0.01 Cofactor metabolic process 5 <0.01 Tetrapyrrole metabolic process 3 <0.01 Porphyrin metabolic process 3 <0.01 Pigment metabolic process 3 <0.01 Annotation cluster 5 (N=8) Enrichment score: 2.15 B-cell translocation gene 3 NM_019290 Glutamate-ammonia ligase M29599 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heme oxygenase 1 NM_012580 Inhibitor of dna binding 2 NM_013060 Peripheral myelin protein 22 NM_017037 Prostaglandin-endoperoxide synthase 2 NM_017232 Timp metallopeptidase inhibitor 2 AJ409332 Term Count (n) Pvalue Regulation of cell proliferation 8 <0.01 Negative regulation of cell proliferation 5 0.01 Regulation of cell cycle 4 0.03 Annotation cluster 6 (N=5) Enrichment score: 1.93 Gamma-glutamyltransferase 5 NM_019235 Glutamate-ammonia ligase M29599 Glutathione s-transferase, theta 2 NM_012796 Solute carrier family 6, member 3 M80233 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Cellular amino acid derivative metabolic process 4 0.01 Cellular amino acid derivative biosynthetic process 3 0.01 Cellular amino acid and derivative metabolic process 5 0.01 Annotation cluster 7 (N=9) Enrichment score: 1.70 Aminolevulinate, delta-, synthase 2 NM_013197 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Inhibitor of dna binding 2 NM_013060 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Mal, t-cell differentiation protein NM_012798 Peripheral myelin protein 22 NM_017037 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Rab9a, member ras oncogene family AF325692 Term Count (n) Pvalue Homeostatic process 9 <0.01 Ion homeostasis 6 <0.01 Chemical homeostasis 6 0.01 Cellular ion homeostasis 5 0.02 Cellular chemical homeostasis 5 0.02 Cellular homeostasis 5 0.04 Annotation cluster 8 (N=12) Enrichment score: 1.63 Aminolevulinate, delta-, synthase 2 NM_013197 Gamma-glutamyltransferase 5 NM_019235 Glutamate-ammonia ligase M29599 Glutathione s-transferase, theta 2 NM_012796 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Hydroxymethylbilane synthase X06827 Polymerase, beta NM_017141 Solute carrier family 6, member 3 M80233 Transcription factor 12 NM_013176 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Amine binding 5 <0.01 Carboxylic acid binding 4 0.01 Nitrogen compound biosynthetic process 5 0.01 Cellular amino acid and derivative metabolic process 5 0.01 Amino acid binding 3 0.02 Amine biosynthetic process 3 0.02 Annotation cluster 9 (N=13) Enrichment score: 1.61 Collagen, type i, alpha 1 M12199 Gamma-glutamyltransferase 5 NM_019235 Glutamate decarboxylase 1 NM_017007 Glutamate-ammonia ligase M29599 Heme oxygenase 1 NM_012580 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Mal, t-cell differentiation protein NM_012798 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Peripheral myelin protein 22 NM_017037 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Solute carrier family 6, member 3 M80233 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Neurotransmitter metabolic process 3 <0.01 Transmission of nerve impulse 5 0.01 Cell communication 6 0.01 Carboxylic acid metabolic process 6 0.01 Oxoacid metabolic process 6 0.01 Organic acid metabolic process 6 0.02 Cellular ketone metabolic process 6 0.02 Regulation of neurotransmitter levels 3 0.02 Annotation cluster 10 (N=21) Enrichment score: 1.59 Adam metallopeptidase with thrombospondin type 1 motif, 1 AF159096 Aminolevulinate, delta-, synthase 2 NM_013197 Collagen, type i, alpha 1 M12199 Glutamate decarboxylase 1 NM_017007 Glutamate-ammonia ligase M29599 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Homer homolog 1 AB007688 Inhibitor of dna binding 2 NM_013060 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Mal, t-cell differentiation protein NM_012798 Peripheral myelin protein 22 NM_017037 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Proline arginine-rich end leucine-rich repeat protein AF163569 Prostaglandin-endoperoxide synthase 2 NM_017232 Rab9a, member ras oncogene family AF325692 Signal-regulatory protein alpha D85183 Solute carrier family 6, member 3 M80233 Timp metallopeptidase inhibitor 2 AJ409332 Transcription factor 12 NM_013176 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Regulation of biological quality 13 <0.01 System process 12 0.02 Developmental process 14 0.02 Multicellular organismal development 13 0.02 System development 12 0.02 Multicellular organismal process 19 0.03 Anatomical structure development 12 0.03 Annotation cluster 11 (N=14) Enrichment score: 1.53 Glutamate-ammonia ligase M29599 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Homer homolog 1 AB007688 Inhibitor of dna binding 2 NM_013060 Lectin, galactoside-binding, soluble, 9 U59462 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Signal-regulatory protein alpha D85183 Solute carrier family 6, member 3 M80233 Timp metallopeptidase inhibitor 2 AJ409332 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Regulation of multicellular organismal process 10 <0.01 Regulation of transport 7 <0.01 Regulation of localization 7 0.01 Regulation of system process 5 0.01 Regulation of calcium ion transport 3 0.01 Regulation of metal ion transport 3 0.02 Regulation of ion transport 3 0.03 Regulation of cell communication 7 0.04 Annotation cluster 12 (N=14) Enrichment score: 1.48 Adam metallopeptidase with thrombospondin type 1 motif, 1 AF159096 Aminolevulinate, delta-, synthase 2 NM_013197 Annexin a3 NM_012823 Glutamate-ammonia ligase M29599 Heme oxygenase 1 NM_012580 Hippocalcin-like 1 NM_017356 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Polymerase, beta NM_017141 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Sparc-like 1 (hevin) NM_012946 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Metal ion binding 14 0.02 Cation binding 14 0.02 Ion binding 14 0.02 Annotation cluster 13 (N=10) Enrichment score: 1.45 B-cell translocation gene 3 NM_019290 Collagen, type i, alpha 1 M12199 Gamma-glutamyltransferase 5 NM_019235 Heme oxygenase 1 NM_012580 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Solute carrier family 6, member 3 M80233 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Response to oxidative stress 5 <0.01 Response to inorganic substance 5 0.01 Response to steroid hormone stimulus 5 0.01 Response to hormone stimulus 6 0.01 Annotation cluster 14 (N=23) Enrichment score: 1.40 3-hydroxy-3-methylglutaryl-coenzyme a synthase 1 X52625 Adam metallopeptidase with thrombospondin type 1 motif, 1 AF159096 Aminolevulinate, delta-, synthase 2 NM_013197 Collagen, type i, alpha 1 M12199 Fucosyltransferase 2 AB006138 Gamma-glutamyltransferase 5 NM_019235 Glutamate decarboxylase 1 NM_017007 Glutamate-ammonia ligase M29599 Glutathione s-transferase, theta 2 NM_012796 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Hydroxymethylbilane synthase X06827 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Mitogen-activated protein kinase 4 Z21935 Polymerase, beta NM_017141 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Rab9a, member ras oncogene family AF325692 Solute carrier family 6, member 3 M80233 Transcription factor 12 NM_013176 Tyrosine hydroxylase NM_012740 Zinc finger protein 36 AB025017 Term Count (n) Pvalue Biosynthetic process 13 0.01 Cellular biosynthetic process 12 0.02 Cellular metabolic process 20 0.03 Metabolic process 22 0.05 Annotation cluster 15 (N=4) Enrichment score: 1.33 Aminolevulinate, delta-, synthase 2 NM_013197 Heme oxygenase 1 NM_012580 Prostaglandin-endoperoxide synthase 2 NM_017232 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Iron ion binding 4 0.04 Heme binding 3 0.05 Annotation cluster 16 (N=7) Enrichment score: 1.33 Aminolevulinate, delta-, synthase 2 NM_013197 Collagen, type i, alpha 1 M12199 Heme oxygenase 1 NM_012580 Malic enzyme 1, nadp(+)-dependent, cytosolic NM_012600 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Response to steroid hormone stimulus 5 0.01 Response to hormone stimulus 6 0.01 Iron ion binding 4 0.04 Oxidoreductase activity 3 0.05 Annotation cluster 17 (N=6) Enrichment score: 1.15 Collagen, type i, alpha 1 M12199 Heme oxygenase 1 NM_012580 Inositol 1,4,5-triphosphate receptor, type 3 NM_013138 Prostaglandin-endoperoxide synthase 2 NM_017232 Solute carrier family 6, member 3 M80233 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Response to inorganic substance 5 0.01 Annotation cluster 18 (N=4) Enrichment score: 1.14 Aminolevulinate, delta-, synthase 2 NM_013197 Heme oxygenase 1 NM_012580 Prostaglandin-endoperoxide synthase 2 NM_017232 Tyrosine hydroxylase NM_012740 Term Count (n) Pvalue Iron ion binding 4 0.04 Annotation cluster 19 (N=8) Enrichment score: 1.06 3-hydroxy-3-methylglutaryl-coenzyme a synthase 1 X52625 B-cell translocation gene 3 NM_019290 Collagen, type i, alpha 1 M12199 Gamma-glutamyltransferase 5 NM_019235 Heat shock 70kd protein 1b; heat shock 70kd protein 1a Z75029 Heme oxygenase 1 NM_012580 Prostaglandin-endoperoxide synthase 2 NM_017232 Solute carrier family 6, member 3 M80233 Term Count (n) Pvalue Response to oxidative stress 5 <0.01 Response to organic nitrogen 3 0.04 Annotation cluster 20 (N=7) Enrichment score: 1.03 Glutamate-ammonia ligase M29599 Guanine nucleotide binding protein, alpha inhibiting 2 M17528 Heme oxygenase 1 NM_012580 Lectin, galactoside-binding, soluble, 9 U59462 Potassium inwardly rectifying channel, subfamily j, member 11 D86039 Prostaglandin-endoperoxide synthase 2 NM_017232 Timp metallopeptidase inhibitor 2 AJ409332 Term Count (n) Pvalue Positive regulation of cell communication 5 0.01 Regulation of cell communication 7 0.04 N, total number of genes in the cluster group. n, total number of genes under the GO term Table 5. DAVID analysis for LVEC – The main effect of AO (79 genes were analysed)
Annotation cluster 1 (N=25) Enrichment score: 2.94 Acetyl-coenzyme a carboxylase alpha X53003 Actin, alpha 1, skeletal muscle V01224 Adam metallopeptidase domain 17 NM_020306 B-cell translocation gene 2, anti-proliferative NM_017259 Carnitine o-octanoyltransferase U26033 Cd47 molecule AF017437 Chemokine ligand 1 D11445 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Chemokine receptor 3 Y13400 Cholinergic receptor, nicotinic, alpha 7 S53987 Endoplasmic reticulum aminopeptidase 1 AF148323 Glutamate receptor, metabotropic 2 M92075 Glycogen synthase kinase 3 beta X53428 Growth hormone receptor U44722 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Guanine nucleotide binding protein, gamma 10 AF022090 Inositol polyphosphate phosphatase-like 1 AB025794 Interleukin 6 M26744 Mechanistic target of rapamycin NM_019906 Metallothionein 1a M11794 Metallothionein 2a M11794 Phosphatidylethanolamine binding protein 2 AF226629 Proteasome (prosome, macropain) subunit, beta type 2 D21799 Similar to nuclear migration protein nudc; nuclear distribution gene c homolog NM_017271 Tumor necrosis factor NM_012675 Term Count (n) Pvalue Response to endogenous stimulus 15 <0.01 Response to organic substance 18 <0.01 Response to hormone stimulus 10 <0.01 Annotation cluster 2 (N=25) Enrichment score: 2.13 5-hydroxytryptamine receptor 3a U59672 Adam metallopeptidase domain 17 NM_020306 Amyloid beta precursor protein-binding, family a, member 3 AF029109 Calcium channel, voltage-dependent, gamma subunit 1 NM_019255 Carnitine o-octanoyltransferase U26033 Cd47 molecule AF017437 Chemokine ligand 1 D11445 Cholinergic receptor, nicotinic, alpha 7 S53987 Clathrin, heavy chain NM_019299 Ferredoxin 1 NM_017126 Glutamate receptor, metabotropic 2 M92075 Glycogen synthase kinase 3 beta X53428 Growth hormone receptor U44722 Interleukin 6 M26744 Mal, t-cell differentiation protein NM_012798 Nck-associated protein 1 D84346 Peptidase beta D13907 Sh3-domain binding protein 4 AJ278266 Similar to hypothetical protein; hypothetical gene supported by x51706; similar to ribosomal protein l9; similar to 60s ribosomal protein l9; ribosomal protein l9; eh-domain containing 2 AF081251 Similar to nuclear migration protein nudc; nuclear distribution gene c homolog NM_017271 Similar to voltage-dependent anion-selective channel protein 2; voltage- dependent anion channel 2 AF268468 Sterol carrier protein 2 M62763 Tenascin xa U24489 Translocase of inner mitochondrial membrane 23 homolog NM_019352 Tumor necrosis factor NM_012675 Term Count (n) Pvalue Localization 25 <0.01 Establishment of localization 22 0.01 Transport 21 0.01 Annotation cluster 3 (N=6) Enrichment score: 1.62 Cd47 molecule AF017437 Cholinergic receptor, nicotinic, alpha 7 S53987 Clathrin, heavy chain NM_019299 Growth hormone receptor U44722 Sh3-domain binding protein 4 AJ278266 Similar to hypothetical protein; hypothetical gene supported by x51706; similar to ribosomal protein l9; similar to 60s ribosomal protein l9; ribosomal protein l9; eh-domain containing 2 AF081251 Term Count (n) Pvalue Membrane invagination 5 0.01 Endocytosis 5 0.01 Membrane organization 6 0.02 Annotation cluster 4 (N=11) Enrichment score: 1.56 Actin, alpha 1, skeletal muscle V01224 Adam metallopeptidase domain 17 NM_020306 Cell division cycle 20 homolog U05341 Cyclin g1 NM_012923 Glycogen synthase kinase 3 beta X53428 Mechanistic target of rapamycin NM_019906 Proteasome subunit, alpha type 4 NM_017281 Proteasome subunit, beta type 2 D21799 Regulator of g-protein signaling 2 AF279918 Similar to nuclear migration protein nudc; nuclear distribution gene c homolog NM_017271 Synaptonemal complex protein 1 NM_012810 Term Count (n) Pvalue Cell cycle process 9 <0.01 Cell cycle 10 <0.01 Mitotic cell cycle 7 <0.01 Cell cycle phase 6 0.01 Cell division 4 0.05 Annotation cluster 5 (N=52) Enrichment score: 1.53 Acetyl-coenzyme a carboxylase alpha X53003 Actin, alpha 1, skeletal muscle V01224 Adam metallopeptidase domain 17 NM_020306 Amyloid beta precursor protein-binding, family a, member 3 AF029109 B-cell translocation gene 2, anti-proliferative NM_017259 Calpastatin Y13588 Ccaat/enhancer binding protein, delta M65149 Cchc-type zinc finger, nucleic acid binding protein D45254 Cd47 molecule AF017437 Cell division cycle 20 homolog U05341 Chemokine ligand 1 D11445 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Chemokine receptor 3 Y13400 Cholinergic receptor, nicotinic, alpha 7 S53987 Cold shock domain protein a U22893 Cyclin g1 NM_012923 Endoplasmic reticulum aminopeptidase 1 AF148323 Eph receptor b1 M59814 Ferredoxin 1 NM_017126 Fibroblast growth factor 1 NM_012846 Fumarate hydratase 1 NM_017005 General transcription factor iia, 2 AF000944 Glutamate receptor, metabotropic 2 M92075 Glycogen synthase kinase 3 beta X53428 Growth hormone receptor U44722 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Guanine nucleotide binding protein, gamma 10 AF022090 Hypocretin receptor 1 NM_013064 Inositol polyphosphate phosphatase-like 1 AB025794 Interleukin 6 M26744 Lactalbumin, alpha NM_012594 Lactate dehydrogenase b NM_012595 Lipopolysaccharide-induced tnf factor U53184 Mal, t-cell differentiation protein NM_012798 Malate dehydrogenase 1, nad AF075574 Mechanistic target of rapamycin NM_019906 Metallothionein 1a M11794 Metallothionein 2a M11794 Nck-associated protein 1 D84346 Nuclear receptor subfamily 1, group d, member 1 M25804 Phosphatidylethanolamine binding protein 2 AF226629 Proteasome subunit, alpha type 4 NM_017281 Proteasome subunit, beta type 2 D21799 Rap guanine nucleotide exchange factor 3 U78167 Regulator of g-protein signaling 2 AF279918 Rho gtpase activating protein 17 AB042827 Similar to nuclear migration protein nudc; nuclear distribution gene c NM_017271 homolog Synaptonemal complex protein 1 NM_012810 Tenascin xa U24489 Tumor necrosis factor NM_012675 Zinc finger protein 36 X63369 Zinc finger, dhhc-type containing 2 AF228917 Term Count (n) Pvalue Regulation of protein modification process 11 <0.01 Positive regulation of protein modification process 9 <0.01 Positive regulation of cellular protein metabolic process 9 <0.01 Positive regulation of protein metabolic process 9 <0.01 Regulation of protein amino acid phosphorylation 8 <0.01 Regulation of protein metabolic process 12 <0.01 Regulation of phosphorylation 11 <0.01 Regulation of phosphate metabolic process 11 <0.01 Regulation of phosphorus metabolic process 11 <0.01 Regulation of catalytic activity 14 <0.01 Regulation of cellular protein metabolic process 11 <0.01 Regulation of protein kinase activity 9 <0.01 Regulation of molecular function 15 <0.01 Interleukin-6 receptor binding 3 <0.01 Positive regulation of protein amino acid phosphorylation 6 <0.01 Regulation of kinase activity 9 <0.01 Regulation of transferase activity 9 <0.01 Positive regulation of phosphorylation 6 <0.01 Positive regulation of phosphorus metabolic process 6 <0.01 Positive regulation of phosphate metabolic process 6 <0.01 Regulation of map kinase activity 6 <0.01 Response to alkaloid 5 <0.01 Positive regulation of chemokine production 3 <0.01 Cell cycle process 9 <0.01 Regulation of signal transduction 13 <0.01 Positive regulation of biological process 22 <0.01 Signal transduction 20 <0.01 Mitotic cell cycle 7 <0.01 Regulation of macromolecule metabolic process 24 <0.01 Regulation of response to external stimulus 6 <0.01 Positive regulation of macromolecule metabolic process 13 <0.01 Response to hormone stimulus 10 <0.01 Regulation of cell communication 14 <0.01 Regulation of protein kinase cascade 7 <0.01 Biological regulation 44 <0.01 Regulation of localization 11 <0.01 Positive regulation of protein kinase cascade 6 <0.01 Positive regulation of cellular metabolic process 13 <0.01 Regulation of chemokine production 3 <0.01 Positive regulation of metabolic process 13 <0.01 Positive regulation of cellular process 19 0.01 Regulation of metabolic process 25 0.01 Positive regulation of signal transduction 7 0.01 Regulation of protein localization 5 0.01 Positive regulation of cell proliferation 8 0.01 Positive regulation of intracellular transport 3 0.01 Positive regulation of molecular function 9 0.01 Response to peptide hormone stimulus 6 0.01 Regulation of biological process 40 0.01 Immune response 8 0.01 Positive regulation of translation 3 0.01 Response to mechanical stimulus 4 0.01 Positive regulation of cell communication 7 0.01 Posttranscriptional regulation of gene expression 5 0.01 Regulation of inflammatory response 4 0.01 Regulation of primary metabolic process 22 0.01 Regulation of response to stimulus 8 0.01 Positive regulation of catalytic activity 8 0.01 Regulation of response to stress 6 0.01 Intracellular signaling cascade 12 0.01 Negative regulation of cytokine production 3 0.02 Receptor binding 10 0.02 Regulation of interleukin-6 production 3 0.02 Glucose metabolic process 5 0.02 Regulation of transmission of nerve impulse 5 0.02 Locomotion 7 0.02 Regulation of cellular metabolic process 22 0.02 Negative regulation of response to stimulus 4 0.02 Regulation of neurological system process 5 0.02 Regulation of multicellular organismal process 12 0.02 Regulation of intracellular protein transport 3 0.02 Hexose metabolic process 5 0.03 Negative regulation of cellular metabolic process 9 0.03 Regulation of mapkkk cascade 4 0.03 Regulation of nucleocytoplasmic transport 3 0.03 Positive regulation of peptidyl-tyrosine phosphorylation 3 0.03 Cell motion 7 0.03 Negative regulation of response to external stimulus 3 0.03 Regulation of cell proliferation 9 0.03 Regulation of system process 6 0.03 Inflammatory response 5 0.03 Positive regulation of protein transport 3 0.04 Cell motility 6 0.04 Localization of cell 6 0.04 Response to insulin stimulus 4 0.04 Response to external stimulus 10 0.04 Negative regulation of apoptosis 6 0.04 Negative regulation of metabolic process 9 0.04 Regulation of developmental process 9 0.04 Regulation of intracellular transport 3 0.04 Monosaccharide metabolic process 5 0.04 Regulation of defense response 4 0.04 Negative regulation of programmed cell death 6 0.04 Negative regulation of cell death 6 0.04 Regulation of anatomical structure morphogenesis 5 0.04 Activation of mapk activity 3 0.05 Cellular carbohydrate metabolic process 6 0.05 Regulation of transport 7 0.05 Generation of precursor metabolites and energy 5 0.05 Immune system process 9 0.05 Cytokine-mediated signaling pathway 3 0.05 Regulation of cellular process 35 0.05 Positive regulation of multicellular organismal process 5 0.05 Annotation cluster 6 (N=8) Enrichment score: 1.45 Adam metallopeptidase domain 17 NM_020306 Cchc-type zinc finger, nucleic acid binding protein D45254 Cd47 molecule AF017437 Cholinergic receptor, nicotinic, alpha 7 S53987 Fibroblast growth factor 1 NM_012846 Interleukin 6 M26744 Mechanistic target of rapamycin NM_019906 Tumor necrosis factor NM_012675 Term Count (n) Pvalue Positive regulation of cell proliferation 8 0.01 Positive regulation of cell adhesion 3 0.05 Annotation cluster 7 (N=27) Enrichment score: 1.37 Adam metallopeptidase domain 17 NM_020306 Calpastatin Y13588 Cathepsin l1 S85184 Cell division cycle 20 homolog U05341 Cholinergic receptor, nicotinic, alpha 7 S53987 Cold shock domain protein a U22893 Cyclin g1 NM_012923 Endoplasmic reticulum aminopeptidase 1 AF148323 Fumarate hydratase 1 NM_017005 Glycogen synthase kinase 3 beta X53428 Growth hormone receptor U44722 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Inositol polyphosphate phosphatase-like 1 AB025794 Interleukin 6 M26744 Lactate dehydrogenase b NM_012595 Lysosomal-associated membrane protein 1 U75406 Malate dehydrogenase 1, nad AF075574 Mechanistic target of rapamycin NM_019906 Peptidase beta D13907 Praja 2, ring-h2 motif containing D32249 Proteasome subunit, alpha type 4 NM_017281 Proteasome subunit, beta type 2 D21799 Proteasome subunit, beta type 7 AF285103 Similar to hypothetical protein; hypothetical gene supported by x51706; AF081251 similar to ribosomal protein l9; similar to 60s ribosomal protein l9; ribosomal protein l9; eh-domain containing 2 Similar to nuclear migration protein nudc; nuclear distribution gene c homolog NM_017271 Tumor necrosis factor NM_012675 Zinc finger protein 36 X63369 Term Count (n) Pvalue Mitotic cell cycle 7 <0.01 Negative regulation of catalytic activity 7 <0.01 Threonine-type endopeptidase activity 3 <0.01 Threonine-type peptidase activity 3 <0.01 Cellular catabolic process 11 0.01 Negative regulation of molecular function 7 0.01 Cellular macromolecule catabolic process 8 0.01 Proteolysis involved in cellular protein catabolic process 7 0.01 Cellular protein catabolic process 7 0.01 Catabolic process 12 0.01 Protein catabolic process 7 0.01 Positive regulation of catalytic activity 8 0.01 Macromolecule catabolic process 8 0.02 Negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.04 Anaphase-promoting complex-dependent proteasomal ubiquitin- dependent protein catabolic process 3 0.04 Negative regulation of ubiquitin-protein ligase activity 3 0.04 Negative regulation of ligase activity 3 0.04 Positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.04 Regulation of ubiquitin-protein ligase activity during mitotic cell cycle 3 0.04 Positive regulation of ubiquitin-protein ligase activity 3 0.04 Endopeptidase activity 6 0.04 Positive regulation of ligase activity 3 0.05 Negative regulation of protein ubiquitination 3 0.05 Regulation of ubiquitin-protein ligase activity 3 0.05 Peptidase activity, acting on l-amino acid peptides 7 0.05 Annotation cluster 8 (N=28) Enrichment score: 1.11 5-hydroxytryptamine (serotonin) receptor 3a U59672 Acetyl-coenzyme a carboxylase alpha X53003 Actin, alpha 1, skeletal muscle V01224 Adam metallopeptidase domain 17 NM_020306 Amyloid beta precursor protein-binding, family a, member 3 AF029109 B-cell translocation gene 2, anti-proliferative NM_017259 Calpastatin Y13588 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Cholinergic receptor, nicotinic, alpha 7 S53987 Cold shock domain protein a U22893 Eph receptor b1 M59814 Fibroblast growth factor 1 NM_012846 Fumarate hydratase 1 NM_017005 Glutamate receptor, metabotropic 2 M92075 Glutamine/glutamic acid-rich protein a M31032 Glycogen synthase kinase 3 beta X53428 Growth hormone receptor U44722 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Hypothetical gene supported by m31032; nm_181440 M31032 Inositol polyphosphate phosphatase-like 1 AB025794 Interferon-related developmental regulator 1 NM_019242 Interleukin 6 M26744 Mal, t-cell differentiation protein NM_012798 Mechanistic target of rapamycin NM_019906 Nck-associated protein 1 D84346 Regulator of g-protein signaling 2 AF279918 Tenascin xa U24489 Tumor necrosis factor NM_012675 Term Count (n) Pvalue Developmental process 23 0.03 Anatomical structure development 20 0.03 Organ development 16 0.04 System development 19 0.04 Multicellular organismal development 20 0.06 Cell differentiation 14 0.07 Anatomical structure morphogenesis 11 0.09 Cellular developmental process 14 0.09 Nervous system development 10 0.10 Generation of neurons 7 0.12 Neurogenesis 7 0.16 Multicellular organismal process 27 0.42 Annotation cluster 9 (N=11) Enrichment score: 1.10 5-hydroxytryptamine receptor 3a U59672 Actin, alpha 1, skeletal muscle V01224 Amyloid beta precursor protein-binding, family a, member 3 AF029109 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Cholinergic receptor, nicotinic, alpha 7 S53987 Eph receptor b1 M59814 Fibroblast growth factor 1 NM_012846 Glutamate receptor, metabotropic 2 M92075 Interleukin 6 M26744 Mal, t-cell differentiation protein NM_012798 Tenascin xa U24489 Term Count (n) Pvalue Cell communication 9 0.01 Cell-cell signaling 7 0.01 Transmission of nerve impulse 6 0.02 Annotation cluster 10 (N=8) Enrichment score: 1.09 Acetyl-coenzyme a carboxylase alpha X53003 Carnitine o-octanoyltransferase U26033 Fumarate hydratase 1 NM_017005 Growth hormone receptor U44722 Inositol polyphosphate phosphatase-like 1 AB025794 Lactate dehydrogenase b NM_012595 Malate dehydrogenase 1, nad AF075574 Tenascin xa U24489 Term Count (n) Pvalue Acetyl-coa metabolic process 3 0.02 Dicarboxylic acid metabolic process 3 0.02 Cellular amide metabolic process 3 0.04 Annotation cluster 11 (N=13) Enrichment score: 1.01 Cathepsin l1 S85184 Chemokine ligand 1 D11445 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Chemokine receptor 3 Y13400 Cholinergic receptor, nicotinic, alpha 7 S53987 Clathrin, heavy chain NM_019299 Eph receptor b1 M59814 Glutamate receptor, metabotropic 2 M92075 Growth hormone receptor U44722 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Guanine nucleotide binding protein, gamma 10 AF022090 Hypocretin receptor 1 NM_013064 Interleukin 6 M26744 Term Count (n) Pvalue Peptide binding 6 <0.01 Peptide hormone binding 3 0.02 Behavior 7 0.03 Hormone binding 3 0.03
N, total number of genes in the cluster group. n, total number of genes under the GO term Table 6. DAVID analysis for LVEC – The main effect of Ex (69 genes were analysed)
Annotation cluster 1 (N=31) Enrichment score: 3.39 Actin, alpha 1, skeletal muscle V01224 Allograft inflammatory factor 1 D82069 Aminolevulinate, delta-, synthase 2 D86297 Atp-binding cassette, sub-family b, member 4 NM_012690 B-cell translocation gene 3 NM_019290 Carnitine o-octanoyltransferase U26033 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Collagen, type xviii, alpha 1 AF189709 Complement factor d S73894 Egl nine homolog 3 NM_019371 Enolase 3, beta, muscle NM_012949 NM_012844, Epoxide hydrolase 1, microsomal M15345 Fatty acid binding protein 3, muscle and heart J02773 Glutathione s-transferase yc2 subunit S82820 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Interleukin 8 receptor, beta NM_017183 Lipoprotein lipase L03294 Nadh dehydrogenase 1 alpha subcomplex 10 X89822 Parkinson disease 7; similar to dj-1 protein AF157511 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Serine peptidase inhibitor, clade h, member 1 NM_017173 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Sphingomyelin phosphodiesterase 2, neutral AB047002 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Vitronectin U44845 Term Count (n) Pvalue Response to drug 16 <0.01 Response to endogenous stimulus 13 <0.01 Response to stress 20 <0.01 Response to organic substance 15 <0.01 Response to chemical stimulus 25 <0.01 Response to steroid hormone stimulus 7 <0.01 Response to hormone stimulus 9 <0.01 Response to stimulus 31 0.01 Response to extracellular stimulus 6 0.02 Response to glucocorticoid stimulus 4 0.03 Response to corticosteroid stimulus 4 0.03 Annotation cluster 2 (N=16) Enrichment score: 2.54 Actin, alpha 1, skeletal muscle V01224 Allograft inflammatory factor 1 D82069 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Cold shock domain protein a U22893 Collagen, type xviii, alpha 1 AF189709 Insulin-like growth factor binding protein 3 NM_012588 Mal, t-cell differentiation protein NM_012798 Peptidylglycine alpha-amidating monooxygenase NM_013000 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Sphingomyelin phosphodiesterase 2, neutral AB047002 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Y box binding protein 1 M69138 Term Count (n) Pvalue Regulation of apoptosis 13 <0.01 Regulation of programmed cell death 13 <0.01 Regulation of cell death 13 <0.01 Regulation of smooth muscle cell proliferation 5 <0.01 Response to abiotic stimulus 9 <0.01 Positive regulation of apoptosis 7 0.01 Positive regulation of programmed cell death 7 0.01 Response to mechanical stimulus 4 0.01 Positive regulation of cell death 7 0.01 Induction of programmed cell death 5 0.02 Induction of apoptosis 5 0.02 Annotation cluster 3 (N=13) Enrichment score: 2.10 Allograft inflammatory factor 1 D82069 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Collagen, type xviii, alpha 1 AF189709 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Insulin-like growth factor binding protein 3 NM_012588 Janus kinase 3 D28508 Lipoprotein lipase L03294 Platelet-derived growth factor alpha polypeptide L06238 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Synaptojanin 2 binding protein AF260258 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue regulation of localization 13 <0.01 Annotation cluster 4 (N=21) Enrichment score: 1.84 Atp-binding cassette, sub-family b, member 4 NM_012690 Carnitine o-octanoyltransferase U26033 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Clathrin, heavy chain NM_019299 Fatty acid binding protein 3, muscle and heart J02773 Fatty acid binding protein 9, testis U07870 Interleukin 8 receptor, beta NM_017183 Mal, t-cell differentiation protein NM_012798 Nadh dehydrogenase 1 alpha subcomplex 10 X89822 Nadh dehydrogenase fe-s protein 6 NM_019223 Parkinson disease 7; similar to dj-1 protein AF157511 Potassium voltage gated channel, shab-related subfamily, member 1 NM_013186 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Solute carrier family 2, member 2 NM_012879 Solute carrier family 25, member 11 U84727 Synaptojanin 2 binding protein AF260258 Translocase of inner mitochondrial membrane 23 homolog NM_019352 Transthyretin M60869 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue Transport 20 0.01 Establishment of localization 20 0.01 Localization 21 0.02 Macromolecule localization 10 0.02 Annotation cluster 5 (N=38) Enrichment score: 1.81 Actin, alpha 1, skeletal muscle V01224 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Allograft inflammatory factor 1 D82069 Aminolevulinate, delta-, synthase 2 D86297 B-cell translocation gene 3 NM_019290 Calpastatin Y13588 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Cold shock domain protein a U22893 Collagen, type xviii, alpha 1 AF189709 Complement factor d S73894 D site of albumin promoter binding protein J03179 Egl nine homolog 3 NM_019371 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Hypocretin receptor 1 AF041244 Insulin-like growth factor binding protein 3 NM_012588 Interleukin 8 receptor, beta NM_017183 Janus kinase 3 D28508 Lipoprotein lipase L03294 Mal, t-cell differentiation protein NM_012798 Max interactor 1 NM_013160 Monoamine oxidase b NM_013198 Nuclear receptor subfamily 1, group d, member 1 M25804 Parkinson disease 7; similar to dj-1 protein AF157511 Platelet-derived growth factor alpha polypeptide L06238 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Regulator of g-protein signaling 2 AF279918 Serpine1 mrna binding protein 1 U21718 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Sphingomyelin phosphodiesterase 2, neutral AB047002 Synaptojanin 2 binding protein AF260258 Transthyretin M60869 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Vitronectin U44845 Y box binding protein 1 M69138 Term Count (n) Pvalue Regulation of cell proliferation 14 <0.01 Positive regulation of biological process 20 <0.01 Positive regulation of cellular process 18 <0.01 Biological regulation 38 0.01 Annotation cluster 6 (N=16) Enrichment score: 1.81 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Allograft inflammatory factor 1 D82069 B-cell translocation gene 3 NM_019290 Carnitine o-octanoyltransferase U26033 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Enolase 3, beta, muscle NM_012949 NM_012844, Epoxide hydrolase 1, microsomal M15345 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Interleukin 8 receptor, beta NM_017183 Parkinson disease 7; similar to dj-1 protein AF157511 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue Response to oxidative stress 6 <0.01 Response to inorganic substance 7 <0.01 Response to hydrogen peroxide 4 0.01 Response to reactive oxygen species 4 0.01 Response to cytokine stimulus 4 0.02 Response to organic nitrogen 4 0.02 Positive regulation of catalytic activity 7 0.03 Aging 4 0.04 Positive regulation of molecular function 7 0.05 Annotation cluster 7 (N=30) Enrichment score: 1.79 Actin, alpha 1, skeletal muscle V01224 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Aminolevulinate, delta-, synthase 2 D86297 Calpastatin Y13588 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Cold shock domain protein a U22893 Collagen, type xviii, alpha 1 AF189709 Enolase 3, beta, muscle NM_012949 Fatty acid binding protein 9, testis U07870 Glutamine/glutamic acid-rich protein a M31032 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Hypothetical gene supported by m31032; nm_181440 M31032 Insulin-like growth factor binding protein 3 NM_012588 Mal, t-cell differentiation protein NM_012798 Monoamine oxidase b NM_013198 Myosin, light polypeptide 2, regulatory, cardiac, slow X07314 Parkinson disease 7; similar to dj-1 protein AF157511 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Regulator of g-protein signaling 2 AF279918 Serine peptidase inhibitor, clade h, member 1 NM_017173 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Tropomodulin 1 NM_013044 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Y box binding protein 1 M69138 Term Count (n) Pvalue Developmental process 24 <0.01 Anatomical structure development 21 0.01 Anatomical structure morphogenesis 13 0.01 Organ development 16 0.02 Multicellular organismal development 20 0.02 System development 18 0.03 Annotation cluster 8 (N=24) Enrichment score: 1.63 Actin, alpha 1, skeletal muscle V01224 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Aminolevulinate, delta-, synthase 2 D86297 Calpastatin Y13588 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Cold shock domain protein a U22893 Collagen, type xviii, alpha 1 AF189709 Enolase 3, beta, muscle NM_012949 Fatty acid binding protein 9, testis U07870 Glutamine/glutamic acid-rich protein a M31032 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Hypothetical gene supported by m31032; nm_181440 M31032 Insulin-like growth factor binding protein 3 NM_012588 Mal, t-cell differentiation protein NM_012798 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Rab3a, member ras oncogene family NM_013018 Regulator of g-protein signaling 2 AF279918 Tropomodulin 1 NM_013044 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Y box binding protein 1 M69138 Term Count (n) Pvalue Developmental process 24 <0.01 Anatomical structure development 21 0.01 Anatomical structure formation involved in morphogenesis 7 0.01 Cellular component assembly involved in morphogenesis 3 0.01 Cell differentiation 14 0.03 Cellular developmental process 14 0.05 Annotation cluster 9 (N=21) Enrichment score: 1.48 Actin, alpha 1, skeletal muscle V01224 Allograft inflammatory factor 1 D82069 Calpastatin Y13588 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Clathrin, heavy chain NM_019299 Collagen, type xviii, alpha 1 AF189709 Fatty acid binding protein 9, testis U07870 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Lipoprotein lipase L03294 Nsfl1 cofactor Y10769 Platelet-derived growth factor alpha polypeptide L06238 Potassium voltage gated channel, shab-related subfamily, member 1 NM_013186 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Serine peptidase inhibitor, clade h, member 1 NM_017173 Synaptojanin 2 binding protein AF260258 Tropomodulin 1 NM_013044 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Vitronectin U44845 Term Count (n) Pvalue Cellular component organization 20 <0.01 Anatomical structure formation involved in morphogenesis 7 0.01 Cellular component assembly 9 0.03 Cellular component biogenesis 9 0.05 Annotation cluster 10 (N=7) Enrichment score: 1.46 Aminolevulinate, delta-, synthase 2 D86297 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Egl nine homolog 3 NM_019371 Fatty acid binding protein 3, muscle and heart J02773 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Tumor protein p53 U90328 Term Count (n) Pvalue Response to oxygen levels 6 <0.01 Response to hypoxia 5 0.02 Carboxylic acid binding 4 0.04 Annotation cluster 11 (N=7) Enrichment score: 1.40 Actin, alpha 1, skeletal muscle V01224 Calpastatin Y13588 Collagen, type xviii, alpha 1 AF189709 Fatty acid binding protein 9, testis U07870 Platelet-derived growth factor alpha polypeptide L06238 Tropomodulin 1 NM_013044 Tumor protein p53 U90328 Term Count (n) Pvalue Anatomical structure formation involved in morphogenesis 7 0.01 Annotation cluster 12 (N=10) Enrichment score: 1.37 Carnitine o-octanoyltransferase U26033 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Clathrin, heavy chain NM_019299 Fatty acid binding protein 3, muscle and heart J02773 Mal, t-cell differentiation protein NM_012798 Rab3a, member ras oncogene family NM_013018 Synaptojanin 2 binding protein AF260258 Translocase of inner mitochondrial membrane 23 homolog NM_019352 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue Intracellular protein transport 7 <0.01 Cellular protein localization 7 0.01 Cellular macromolecule localization 7 0.01 Protein transport 8 0.02 Establishment of protein localization 8 0.02 Macromolecule localization 10 0.02 Intracellular transport 7 0.04 Protein localization 8 0.05 Annotation cluster 13 (N=10) Enrichment score: 1.17 Allograft inflammatory factor 1 D82069 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Complement factor d S73894 Enolase 3, beta, muscle NM_012949 Interleukin 8 receptor, beta NM_017183 Platelet-derived growth factor alpha polypeptide L06238 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Vitronectin U44845 Term Count (n) Pvalue Response to wounding 7 0.02 Humoral immune response 3 0.02 Annotation cluster 14 (N=14) Enrichment score: 1.17 Allograft inflammatory factor 1 D82069 Aminolevulinate, delta-, synthase 2 D86297 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Complement factor d S73894 Insulin-like growth factor binding protein 3 NM_012588 Interleukin 8 receptor, beta NM_017183 Janus kinase 3 D28508 Lipoprotein lipase L03294 Platelet-derived growth factor alpha polypeptide L06238 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Vitronectin U44845 Term Count (n) Pvalue Hemopoiesis 5 0.03 Immune system process 9 0.03 Cell activation 5 0.04 Hemopoietic or lymphoid organ development 5 0.04 Cell proliferation 5 0.04 Immune system development 5 0.05 Annotation cluster 15 (N=13) Enrichment score: 1.17 Actin, alpha 1, skeletal muscle V01224 Aminolevulinate, delta-, synthase 2 D86297 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Janus kinase 3 D28508 Lipoprotein lipase L03294 Mal, t-cell differentiation protein NM_012798 Monoamine oxidase b NM_013198 Parkinson disease 7; similar to dj-1 protein AF157511 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Transthyretin M60869 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue Cellular ion homeostasis 7 0.01 Cellular chemical homeostasis 7 0.01 Chemical homeostasis 8 0.01 Ion homeostasis 7 0.02 Cellular homeostasis 7 0.03 Regulation of biological quality 13 0.04 Annotation cluster 16 (N=10) Enrichment score: 1.06 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Allograft inflammatory factor 1 D82069 Calpastatin Y13588 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Egl nine homolog 3 NM_019371 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Interleukin 8 receptor, beta NM_017183 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Term Count (n) Pvalue Positive regulation of hydrolase activity 5 0.01 Positive regulation of catalytic activity 7 0.03 Cell activation 5 0.04 Positive regulation of molecular function 7 0.05 Regulation of hydrolase activity 5 0.05 Annotation cluster 17 (N=51) Enrichment score: 1.01 Actin, alpha 1, skeletal muscle V01224 Adenylate cyclase activating polypeptide 1 receptor type i D16465 Allograft inflammatory factor 1 D82069 Aminolevulinate, delta-, synthase 2 D86297 B-cell translocation gene 3 NM_019290 Calpastatin Y13588 Carnitine o-octanoyltransferase U26033 Cathepsin l1 S85184 Cd74 molecule, major histocompatibility complex, class ii invariant chain X13044 Cell division cycle 2, g1 to s and g2 to m NM_019296 Chemokine receptor 2; angiotensin ii receptor, type 1b M90065 Coenzyme q3 homolog, methyltransferase NM_019187 Cold shock domain protein a U22893 Collagen, type xviii, alpha 1 AF189709 Complement factor d S73894 D site of albumin promoter binding protein J03179 Egl nine homolog 3 NM_019371 Enolase 3, beta, muscle NM_012949 NM_012844, Epoxide hydrolase 1, microsomal M15345 Farnesyl diphosphate synthase; similar to testis-specific farnesyl pyrophosphate synthetase M34477 Fatty acid binding protein 3, muscle and heart J02773 Glutathione s-transferase yc2 subunit S82820 Guanine nucleotide binding protein, alpha activating activity polypeptide o NM_017327 Insulin-like growth factor binding protein 3 NM_012588 Interleukin 8 receptor, beta NM_017183 Janus kinase 3 D28508 Lactalbumin, alpha NM_012594 Lipoprotein lipase L03294 Mal, t-cell differentiation protein NM_012798 Malate dehydrogenase 1, nad AF075574 Max interactor 1 NM_013160 Monoamine oxidase b NM_013198 Nadh dehydrogenase 1 alpha subcomplex 10 X89822 Nadh dehydrogenase fe-s protein 6 NM_019223 Nuclear receptor subfamily 1, group d, member 1 M25804 Parkinson disease 7; similar to dj-1 protein AF157511 Peptidylglycine alpha-amidating monooxygenase NM_013000 Platelet-derived growth factor alpha polypeptide L06238 Praja 2, ring-h2 motif containing D32249 Purinergic receptor p2x, ligand-gated ion channel 4 U47031 Rab3a, member ras oncogene family NM_013018 Regulator of g-protein signaling 2 AF279918 Serine peptidase inhibitor, clade h, member 1 NM_017173 Serpine1 mrna binding protein 1 U21718 Signal transducer and activator of transcription 1; signal transducer and activator of transcription 4 AF205604 Sphingomyelin phosphodiesterase 2, neutral AB047002 Transthyretin M60869 Tumor necrosis factor NM_012675 Tumor protein p53 U90328 Udp-n-acetyl-alpha-d-galactosamine:polypeptide n- acetylgalactosaminyltransferase 1 U35890 Y box binding protein 1 M69138 Term Count (n) Pvalue Regulation of cell proliferation 14 <0.01 Regulation of apoptosis 13 <0.01 Regulation of programmed cell death 13 <0.01 Regulation of cell death 13 <0.01 Regulation of smooth muscle cell proliferation 5 <0.01 Positive regulation of cell proliferation 9 <0.01 Regulation of cell migration 6 <0.01 Regulation of cell motion 6 <0.01 Regulation of locomotion 6 <0.01 Negative regulation of apoptosis 7 0.01 Negative regulation of programmed cell death 7 0.01 Negative regulation of cell death 7 0.01 Positive regulation of cell migration 4 0.01 Regulation of protein modification process 6 0.01 Positive regulation of cell motion 4 0.01 Positive regulation of locomotion 4 0.02 Regulation of mitotic cell cycle 4 0.02 Single-stranded dna binding 3 0.02 Response to extracellular stimulus 6 0.02 Positive regulation of smooth muscle cell proliferation 3 0.02 Regulation of striated muscle tissue development 3 0.02 Response to wounding 7 0.02 Regulation of muscle development 3 0.02 Positive regulation of catalytic activity 7 0.03 Regulation of multicellular organismal process 11 0.03 Negative regulation of transcription from rna polymerase ii promoter 5 0.03 Biosynthetic process 18 0.03 Regulation of protein metabolic process 7 0.04 Cell activation 5 0.04 Positive regulation of cell cycle 3 0.04 Cell proliferation 5 0.04 Positive regulation of molecular function 7 0.05
N, total number of genes in the cluster group. n, total number of genes under the GO term Table 7. DAVID analysis for LVEC – The main interaction effects (75 genes were analysed) Annotation cluster 1 (N=61) Enrichment Score: 4.19 Adenosylmethionine decarboxylase 1 M34464 ADP-ribosylarginine hydrolase M86341 ADP-ribosyltransferase 2b M85193 Aldehyde dehydrogenase 7 family, member A1 S75019 Aldo-keto reductase family 1, member B1; aldo-keto reductase family 1, member B1, pseudogene 2 NM_012498 Atpase, Na+/K+ transporting, alpha 2 polypeptide NM_012505 Calbindin 1 M31178 Coagulation factor II receptor NM_012950 Core-binding factor, beta subunit AF087437 D site of albumin promoter binding protein J03179 Dynein cytoplasmic 1 heavy chain 1 NM_019226 Early growth response 1 NM_012551 Epidermal growth factor X12748 Epithelial membrane protein 3 Y10889 Eukaryotic translation initiation factor 2B, subunit 4 delta Z48225 Farnesyl diphosphate synthase; similar to testis-specific farnesyl pyrophosphate synthetase M34477 General transcription factor IIA, 2 AF000944 General transcription factor IIF, polypeptide 2 D10665 Glial fibrillary acidic protein NM_017009 Glutathione S-transferase A2 NM_017013 Growth hormone receptor U44722 Heat shock protein 5 M14050 HMG-box transcription factor 1 NM_013221 Janus kinase 3 NM_012855 Lactate dehydrogenase A; similar to L-lactate dehydrogenase A chain M54926 Lectin, galactoside-binding, soluble, 1 NM_019904 Lipoprotein lipase L03294 Lysosomal-associated membrane protein 1 U75406 Lysozyme 2; lysozyme C type 2 L12458 Mannosidase, alpha, class 2C, member 1 M57547 Milk fat globule-EGF factor 8 protein D84068 Myosin, heavy chain 6, cardiac muscle, alpha NM_017239 Myosin, light chain 6, alkali, smooth muscle and non-muscle; similar to Myosin light chain 1 slow a; myosin, light polypeptide 6, alkali, smooth muscle and non-muscle-like; myosin, light chain 6B, alkali, smooth muscle and non-muscle S77858 NADH dehydrogenase 1 alpha subcomplex 10 X89822 NCK-associated protein 1 D84346 Y10769, NSFL1 cofactor AB002086 Nuclear receptor subfamily 1, group D, member 1 M25804 Parkinson disease 7; similar to DJ-1 protein AF157511 Peroxiredoxin 5 AF110732 Praja 2, RING-H2 motif containing D32249 Proprotein convertase subtilisin/kexin type 6 NM_012999 Proteasome 26S subunit, atpase, 1 D50696 Proteasome activator subunit 2 NM_017257 Protein phosphatase 2, regulatory subunit B, alpha isoform M83298 Protein tyrosine phosphatase, receptor type, A NM_012763 Purinergic receptor P2X, ligand-gated ion channel 4 U47031 Regulator of G-protein signaling 2 AF279918 Ribosomal protein L13A X68282 Ribosomal protein L23; hypothetical gene supported by X58200 X58200 Selenoprotein P, plasma, 1 NM_019192 Similar to 40S ribosomal protein S2; ribosomal protein S2 U92696 Similar to Adenosylhomocysteinase; S-adenosylhomocysteine hydrolase NM_017201 Similar to Proteasome subunit beta type 3 D21800 Splicing factor, arginine/serine-rich 10 D49708 Splicing factor, arginine/serine-rich 12 NM_020092 N, total number of genes in the cluster group. n, total number of genes under the GO term