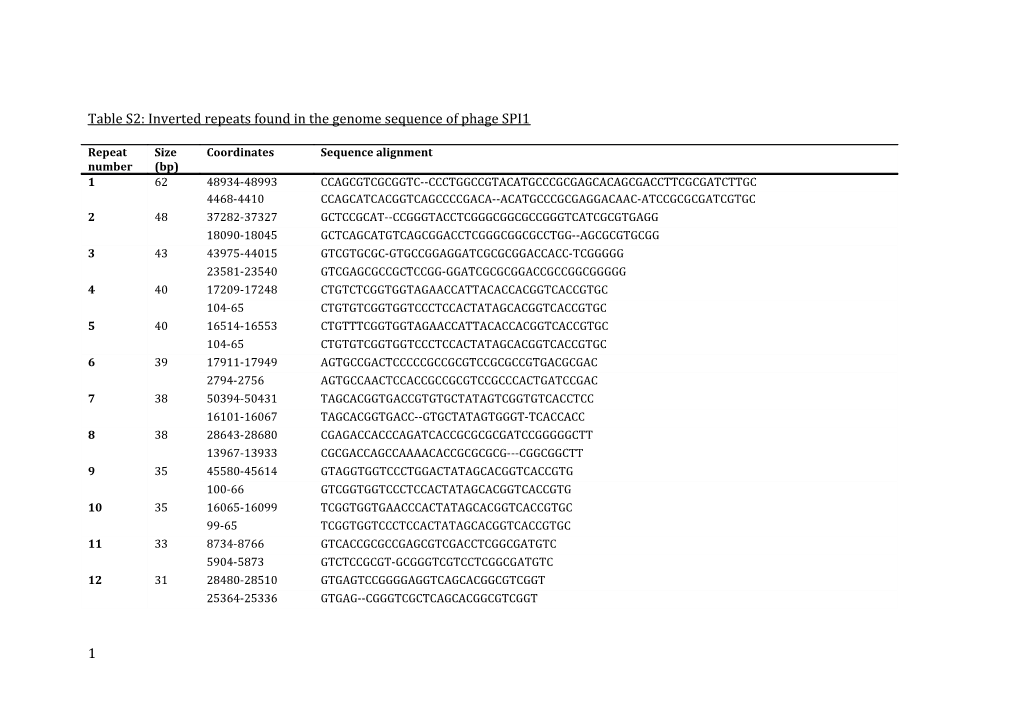
Table S2: Inverted repeats found in the genome sequence of phage SPI1
Repeat Size Coordinates Sequence alignment number (bp) 1 62 48934-48993 CCAGCGTCGCGGTC--CCCTGGCCGTACATGCCCGCGAGCACAGCGACCTTCGCGATCTTGC 4468-4410 CCAGCATCACGGTCAGCCCCGACA--ACATGCCCGCGAGGACAAC-ATCCGCGCGATCGTGC 2 48 37282-37327 GCTCCGCAT--CCGGGTACCTCGGGCGGCGCCGGGTCATCGCGTGAGG 18090-18045 GCTCAGCATGTCAGCGGACCTCGGGCGGCGCCTGG--AGCGCGTGCGG 3 43 43975-44015 GTCGTGCGC-GTGCCGGAGGATCGCGCGGACCACC-TCGGGGG 23581-23540 GTCGAGCGCCGCTCCGG-GGATCGCGCGGACCGCCGGCGGGGG 4 40 17209-17248 CTGTCTCGGTGGTAGAACCATTACACCACGGTCACCGTGC 104-65 CTGTGTCGGTGGTCCCTCCACTATAGCACGGTCACCGTGC 5 40 16514-16553 CTGTTTCGGTGGTAGAACCATTACACCACGGTCACCGTGC 104-65 CTGTGTCGGTGGTCCCTCCACTATAGCACGGTCACCGTGC 6 39 17911-17949 AGTGCCGACTCCCCCGCCGCGTCCGCGCCGTGACGCGAC 2794-2756 AGTGCCAACTCCACCGCCGCGTCCGCCCACTGATCCGAC 7 38 50394-50431 TAGCACGGTGACCGTGTGCTATAGTCGGTGTCACCTCC 16101-16067 TAGCACGGTGACC--GTGCTATAGTGGGT-TCACCACC 8 38 28643-28680 CGAGACCACCCAGATCACCGCGCGCGATCCGGGGGCTT 13967-13933 CGCGACCAGCCAAAACACCGCGCGCG---CGGCGGCTT 9 35 45580-45614 GTAGGTGGTCCCTGGACTATAGCACGGTCACCGTG 100-66 GTCGGTGGTCCCTCCACTATAGCACGGTCACCGTG 10 35 16065-16099 TCGGTGGTGAACCCACTATAGCACGGTCACCGTGC 99-65 TCGGTGGTCCCTCCACTATAGCACGGTCACCGTGC 11 33 8734-8766 GTCACCGCGCCGAGCGTCGACCTCGGCGATGTC 5904-5873 GTCTCCGCGT-GCGGGTCGTCCTCGGCGATGTC 12 31 28480-28510 GTGAGTCCGGGGAGGTCAGCACGGCGTCGGT 25364-25336 GTGAG--CGGGTCGCTCAGCACGGCGTCGGT
1 Repeat Size Coordinates Sequence alignment number (bp) 13 30 54268-54297 CACTATAGCACGGTGACCGTGCTATAGTGG 16106-16077 CACTATAGCACGGTGACCGTGCTATAGTGG 14 30 18628-18657 GCCTACCGTGACGATCGAGGGACAGGTGCC 9330-9301 GCCTACCGTGACGAGCGCGAAGCAGGTACC 15 29 54261-54289 GTCCCTCCACTATAGCACGGTGACCGTGC 93-65 GTCCCTCCACTATAGCACGGTCACCGTGC 16 29 29256-29284 GCGTTCGCGGTGCACACCGCGCGGGCGTC 26940-26912 GCGTTCGCGGTGCCGGCCGCGCCGGGGTC 17 28 54276-54303 CACGGTGACCGTGCTATAGTGGAGGGAC 45614-45587 CACGGTGACCGTGCTATAGTCCAGGGAC 18 27 51018-51044 CACGGTGACCGTGCTATGGTGGAGGGA 45614-45588 CACGGTGACCGTGCTATAGTCCAGGGA 19 27 29173-29199 TCCGCGACGAGCTGGTCGTGCCCGCCG 28332-28306 TCCGCGCCCACGTGGTCGTGCCCGCCG 20 27 49458-49481 GCGACCA---CGTCCGCGCCGACCCCG 8963-8937 GCGACCAGTGCGTCCGCGCCGACCTCG 21 26 44702-44727 CCGGGTCGGTGACCGCGACGATGTCG 11683-11658 CCGGGATGGTGACCGCGAGGCTGTCG 22 26 52813-52838 CTGCCCGCGATCGCGAAGAGACTGGA 49978-49953 CTGGCCGAGATCGCGAAGATCCTGGA 23 25 54273-54297 TAGCACGGTGACCGTGCTATAGTGG 16555-16531 TAGCACGGTGACCGTGGTGTAATGG 24 25 54273-54297 TAGCACGGTGACCGTGCTATAGTGG 17250-17226 TAGCACGGTGACCGTGGTGTAATGG 25 25 23677-23700 ATCG-GCGGCGGTCGATCTCGGGTC 6521-6497 ATCGCGCGGCGGTCGATGTCGGATC
2 Repeat Size Coordinates Sequence alignment number (bp) 26 25 54831-54855 CGCTGGAGGTGTGCGACCTCCGCGA 15522-15498 CGCTGGCGGTCATCGACCTCCGCGA 27 25 35406-35430 CGGCGTGCCGACCGCCGAGTCGACC 24937-24913 CGCCGCGCCGACCGCCGCGCCGACC 28 24 51016-51039 AGCACGGTGACCGTGCTATGGTGG 16100-16077 AGCACGGTGACCGTGCTATAGTGG 29 24 54267-54290 CCACTATAGCACGGTGACCGTGCT 32080-32057 CCACCATAGCACGGTGACCGTGCT 30 24 45599-45622 TAGCACGGTCACCGTGTCAGTCGC 4298-4275 TAGCACGGTCACCGTGCTACTCGC 31 24 27812-27835 GCAGCGGATAGCGGCGGACCGTCC 7203-7180 GCGGCGGTCAGCGGCGGACCGTCC 32 24 41462-41485 CACCCACTCGGGCGCGATCGTCAG 14715-14692 CACCCACTCGGTCCCGAACGTCAG 33 23 17128-17150 CGGACAGGTCGCGCTCGTCGAGC 7939-7917 CGGACCGGACGCGCTCGTCGAGC 34 23 45592-45614 TGGACTATAGCACGGTCACCGTG 16108-16086 TGCACTATAGCACGGTGACCGTG 35 23 17676-17698 TCGATCGCGTCCGTCACCATCGA 1807-1785 TCGATCGCGTCCGCCATCAGCGA 36 23 6500-6522 CCGACATCGACCGCCGCGCGATT 2905-2883 CCGCCCTCGACCGCCGCGCGCTT 37 23 19372-19394 CGTAGCGCACCGACCGCGTAGCC 3842-3820 CGTAGCGCACCGCGCGGGTAGCC 38 22 19163-19184 CACGGTCACCGTGGATGTCCCG 78-57 CACGGTCACCGTGCATTTCCCG
3 Repeat Size Coordinates Sequence alignment number (bp) 39 22 27584-27605 GTCGACGTACGTGGCCCCGGGG 9821-9800 GTAGACGTAGGTGGCCCCGGGG 40 20 41788-41807 GATCGGCGACGATCCCGGCC 533-514 GATCGGCGACGCACCCGGCC 41 20 12230-12249 GACACCGGGGACACGTCGCG 5403-5384 GACACCACGGACACGTCGCG 42 20 18675-18694 GCGGTGCGCGGCGGACCGTC 7200-7181 GCGGTCAGCGGCGGACCGTC 43 20 27206-27225 GTGTGCGCGTGATCGAGCGG 7317-7298 GTGTGCGCGTGCTCGATCGG 44 20 39143-39162 GGCCGCACCCTCACCGACGC 16750-16731 GGGCGCACCCTCACCAACGC 45 20 27509-27528 GTCGTCGGGACCGATCTCGG 18462-18443 GTCGTCGATACCGATCTCGG 46 20 34134-34153 GCAAGCTGACCGTGCTGAAG 32072-32053 GCACGGTGACCGTGCTGAAG 47 19 16083-16101 TAGCACGGTCACCGTGCTA 4298-4280 TAGCACGGTCACCGTGCTA 48 19 23858-23876 GCGGGTGTCGGCGCGGCGG 6402-6384 GCGGGTGTCGGAGCGGCGG 49 19 40821-40839 CGCGAGCGCCGCCGACGCG 24481-24463 CGCGAGCGCCGCCGTCGCG 50 18 32057-32074 AGCACGGTCACCGTGCTA 4297-4280 AGCACGGTCACCGTGCTA 51 18 3439-3456 CTGAGCGCCGACCTCGGC 385-368 CTGGGCGCCGACCTCGGC
4 Repeat Size Coordinates Sequence alignment number (bp) 52 18 12654-12671 CGCGACGACCGGCGCCGC 3119-3102 CGCGACGACCGCCGCCGC 53 18 25657-25674 GTGCGGTACGGGATCACG 9862-9845 GTGCGGTACGGGGTCACG 54 18 32618-32635 CCGATCGGTCGACGCCGG 16330-16313 CCGATCGGTCGATGCCGG 55 18 25924-25941 CCGGTCAGCTCGGCGGTC 18705-18688 CCGGTCAGCTCGACGGTC 56 18 49827-49844 CCGGCCGCGATCGCGGCG 21825-21808 CCGTCCGCGATCGCGGCG 57 18 53929-53946 ACCCGGACGCGGTGGTGG 40314-40297 ACCGGGACGCGGTGGTGG 58 18 55574-55591 CTCGCGGACCTCCACGCG 43623-43606 CTCGCGGACCTCGACGCG 59 17 50393-50409 CTAGCACGGTGACCGTG 16556-16540 CTAGCACGGTGACCGTG 60 17 50393-50409 CTAGCACGGTGACCGTG 17251-17235 CTAGCACGGTGACCGTG 61 17 51016-51032 AGCACGGTGACCGTGCT 32073-32057 AGCACGGTGACCGTGCT 62 17 53414-53430 GATCGGCGCGGTGTCGA 38542-38526 GATCGGCGCGGTGTCGA 63 16 32057-32072 AGCACGGTCACCGTGC 80-65 AGCACGGTCACCGTGC 64 16 17235-17250 CACGGTCACCGTGCTA 4295-4280 CACGGTCACCGTGCTA
5 Repeat Size Coordinates Sequence alignment number (bp) 65 16 16540-16555 CACGGTCACCGTGCTA 4295-4280 CACGGTCACCGTGCTA 66 54424-54439 GTACCGCGCGGTACGA 10481-10466 GTACCGCGCGGTACGA 67 16 36762-36777 TGAGCGGGTCGTCTCC 26212-26197 TGAGCGGGTCGTCTCC 68 16 50394-50409 TAGCACGGTGACCGTG 32074-32059 TAGCACGGTGACCGTG 69 16 53973-53988 GACGACGACGACGACG 46751-46736 GACGACGACGACGACG 70 16 53973-53988 GACGACGACGACGACG 46754-46739 GACGACGACGACGACG 71 16 53973-53988 GACGACGACGACGACG 46757-46742 GACGACGACGACGACG 72 15 50214-50228 TCGGCCGGCATCTCG 10073-10059 TCGGCCGGCATCTCG 73 15 51632-51646 ACCTCCCCCGCCGCG 15846-15832 ACCTCCCCCGCCGCG 74 15 51016-51030 AGCACGGTGACCGTG 16554-16540 AGCACGGTGACCGTG 75 15 37939-37953 CGGCCGCCGCGTCGG 16803-16789 CGGCCGCCGCGTCGG 76 15 51016-51030 AGCACGGTGACCGTG 17249-17235 AGCACGGTGACCGTG 77 15 34584-34598 GCCGATCCGCCACCG 18185-18171 GCCGATCCGCCACCG
6 Repeat Size Coordinates Sequence alignment number (bp) 78 15 47159-47173 CGTCCTCGCTGTACC 26498-26484 CGTCCTCGCTGTACC 79 15 44088-44102 CGAGATCCGGGATCG 37378-37364 CGAGATCCGGGATCG 80 15 51164-51178 CTGCGCGATATCGCC 45146-45132 CTGCGCGATATCGCC
7