Lecture 1: Introduction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions
Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Synthetic Biology: DNA Digital Storage, Computation and the Organic Computer Alex Widdel University of Minnesota, Morris 1 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Outline 1 Overview of Synthetic Biology 2 Background: Systems Architecture and Biology 3 Memory: DNA Storage 4 Building a Biological CPU 5 Conclusions 2 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions What is Synthetic Biology? Biology is viewed as technology. One central goal: construction of a universal bio-computer. A union of biology, computer science, and engineering. The interdisciplinary nature and youth of synthetic biology has led to debate over the term. 3 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Background: Systems Architecture 4 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Systems: Von Neumann Architecture Four parts: Memory Input/output device (IO) Control Unit Arithmetic Logic Unit (ALU) I Arithmetic I/O Logic O Memory Control 5 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Background: Biology 6 / 27 Overview of Synthetic Biology Background: Systems Architecture and Biology Memory: DNA Storage Building a Biological CPU Conclusions Biology: DNA and Protein Synthesis Structure of DNA: DNA can encode bits in a way that is compatible with the way computers store information. -

February Newsletter
Systematic Innovation e-zine Issue 132, March 2013 In this month’s issue: Article – Ten Innovation Trilemmas Article – Holes Not So Funny – Killer Flapjacks Patent of the Month – Temperature Responsive Textiles Best of The Month – Yes To The Mess Conference Report – Hargraves 2013, Sydney Investments – Biological Transistor Generational Cycles – The Wise Prophet Biology – Silver Ant II Short Thort News The Systematic Innovation e-zine is a monthly, subscription only, publication. Each month will feature articles and features aimed at advancing the state of the art in TRIZ and related problem solving methodologies. Our guarantee to the subscriber is that the material featured in the e-zine will not be published elsewhere for a period of at least 6 months after a new issue is released. Readers’ comments and inputs are always welcome. Send them to [email protected] 2013, DLMann, all rights reserved Ten Innovation Trilemmas One day in the not too distant future, we’re hoping to finish the third part in our series on trilemmas. We see the trilemma as the next logical evolution stage to the dilemma (contradiction) story that presently sits at the heart of TRIZ and SI. The basic theory goes something like this: it is relatively easy to resolve a conflict between a pair of system attributes because there is an implied freedom to adversely affect any third attribute that wasn’t part of the problem definition. Solving contradictions helps deliver ‘more ideal’ solutions only in so far as there is an expectation that the two conflicting parameters under consideration are more important than any of the others that might be present in or around the system. -

The Biological Microprocessor, Or How to Build a Computer with Biological Parts
Volume No: 7, Issue: 8, April 2013, e201304003, http://dx.doi.org/10.5936/csbj.201304003 CSBJ The biological microprocessor, or how to build a computer with biological parts Gerd HG Moe-Behrens a,* Abstract: Systemics, a revolutionary paradigm shift in scientific thinking, with applications in systems biology, and synthetic biology, have led to the idea of using silicon computers and their engineering principles as a blueprint for the engineering of a similar machine made from biological parts. Here we describe these building blocks and how they can be assembled to a general purpose computer system, a biological microprocessor. Such a system consists of biological parts building an input / output device, an arithmetic logic unit, a control unit, memory, and wires (busses) to interconnect these components. A biocomputer can be used to monitor and control a biological system. Introduction Nature and computers are words that used to mean unrelated Other important contributions to systemics are by the Nobel-prize things. However, this view changed, starting in the 1940s, when a winning work of Ilya Prigogine on self-organization and his systems revolutionary scientific paradigm, systemics based on platonic theory concepts in thermodynamics [10]. Furthermore: Mitchell idealistic philosophy, gained popularity [1] [2] [3]. Feigenbaums work on Chaos theory [11]. Contemporary application The roots of philosophical idealism based systemics goes back to finds systems theory in bioscience in fields such as systems biology, Plato. A centerpiece of Plato’s (428/7 to 348/7 BC) work is his and its practical application synthetic biology [12]. The term systems theory of forms, also called theory of ideas [2]. -

This New Discovery Will Finally Allow Us to Build Biological Computers
This new discovery will finally allow us to build biological computers The dawn of biological computers is at hand. In a major first for synthetic biology, Stanford engineers have used genetic material to create a biological transistor. Called the "transcriptor," the creation is the final, missing component necessary for the creation of a biological computer that could enable researchers to program functions into living cells. Modern computers rely on three standard functions. One: they must be able to store information. Two: they have to be able to transmit information. Three: they need a basic system of logic – a set of rules that governs how they should function given one or more forms of input. A biological computer would implement all three on a cellular level, using proteins and DNA in place of silicon chips. The first two functions have been demonstrated with cellular materials before. Several labs have now demonstrated the ability to store digital data in DNA,some of them at jaw-dropping densities; and last year, a team led by Stanford bioengineer Drew Endy developed a system for transmitting genetic information between cells. Now, in a study recounted in the latest issue ofScience, Endy's team has developed what it calls a "transcriptor" – the biological equivalent of a digital transistor – and with it a system of logic that can control cellular function. Soon you'll be backing up your hard drive using DNA Think the memory card in your camera is high-capacity? It's got nothing on DNA. With data…Read more io9.com In your standard computer, transistors govern the flow of electricity by playing red light/green light with electrons along a circuit. -

DNA Computing
DNA Computing Uladzislau Maiseichykau Michael Newell Overview ● Introduction to DNA computing ● History ● Differences between DNA and traditional computing ● Potential ● Limitations ● Constructing a DNA computing experiment ● Early DNA computing ● Modern DNA computing ● Future of DNA computing What is DNA Computing? DNA computing uses DNA, chemistry, and molecular hardware instead of traditional silicon-based circuits to run calculations and computations Interest in DNA computing stems from the possibility of processing larger sets of data in a smaller space than silicon computers, and the fast-approaching physical limits to Moore’s Law History ● Initially developed by Leonard Adleman from University of Southern California in 1994 ● Used DNA to solve the Traveling Salesman problem ● In 1997, researchers from University of Rochester used DNA to create logic gates ● Researchers from Weizmann Institute of Science unveiled a programmable computing machine that used enzymes and DNA in 2002 ● 2013, the first biological transistor, the transcriptor, was created Comparison to Standard Computing “DNA computers are unlikely to become stand-alone competitors for electronic computers.” Smallest most efficient computers, modifies cells Build items vs. raw mathematics Potential ● Ultra small devices ● Self assembly (nano) - ex: Sierpinski triangle ● Exotic and new molecular structures ● Nano-Gold + DNA = 10 smaller details on chips Limitations ● Human involvement to set up each experiment/calculation ● Response times can be as slow as minutes, hours, -

DNA Computers
Volume IV, Issue XII, December 2015 IJLTEMAS ISSN 2278 – 2540 DNA Computers Ani Mehrotra Nirma University, India Abstract – The paper discusses the concept of DNA computing, its Although DNA computing is in its infancy, and its advantages over silicon-based computing and the future it holds implications are only beginning to be explored, they have for the mankind. It showcases the limitations of present already been harnessed to perform complex mathematical technology and the promises DNA computing hold for the future. problems.[9] The paper looks on the future we shall be living in and how this new technology can sweep away all the notions of how we II. WHAT IT IS? perceive and live in our world. Keywords – DNA, nucleotides, parallel computing, DNA bases, In its simplest sense, a computer is just a machine capable Hamiltonian Path Problem, Transcriptor, nucleic acids. of performing computations. It doesn't have to be electronic. Tom Ran, of the Weizmann Institute of Science in Israel, I. INTRODUCTION works with computers made out of strands of DNA."Working this way, we can get three trillion computers, working in omputer chip manufacturers are furiously racing to make parallel, in a space the size of a water droplet," he says. The 0s C the next microprocessor that will topple speed records. and 1s of conventional computers are replaced with the four Sooner or later, though, this competition is bound to hit a DNA bases: A, C, G and T. Operations can be translated into wall. Microprocessors made of silicon will eventually reach strands of DNA using these bases, and the way the DNA their limits of speed and miniaturization. -
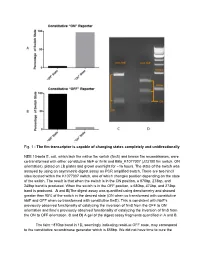
Fig. 1Анаthe Fim Transcriptor Is Capable of Changing
Fig. 1 The fim transcriptor is capable of changing states completely and unidirectionally NEB 10beta E. coli, which lack the native fim switch (fimS) and known fim recombinases, were cotransformed with either constitutive hbiF or fimE and BBa_K1077007 (J23100 fim switch, ON orientation), plated on LB plates and grown overnight for ~16 hours. The state of the switch was assayed by using an asymmetric digest assay on PCR amplified switch. There are two hincII sites located within the K1077007 switch, one of which changes position depending on the state of the switch. The result is that when the switch is in the ON position, a 870bp, 273bp, and 248bp band is produced. When the switch is in the OFF position, a 680bp, 473bp, and 273bp band is produced. A and B)The digest assay was quantified using densitometry and showed greater than 95% of the switch in the desired state (ON when co transformed with constitutive hbiF and OFF when co transformed with constitutive fimE). This is consistent with hbiF’s previously observed functionality of catalyzing the inversion of fimS from the OFF to ON orientation and fime’s previously observed functionality of catalyzing the inversion of fimS from the ON to OFF orientation. C and D) A gel of the digest assay fragments quantified in A and B. The faint ~870bp band in 1D, seemingly indicating residual OFF state, may correspond to the constitutive recombinase generator which is 858bp. We did not have time to cure the bacteria of the constitutive recombinase generator plasmid. Additionally, the switch is on the high copy pSB1C3 plasmid and so it could be that some switch plasmids are escaping recombination. -

Use Style: Paper Title
South Asian Journal of Engineering and Technology Vol.2, No.20 (2016) 1–6 DNA Logic Gates to Design a 4*4 bit Carry Select Adder Circuit Seeja V M Daniel Raj A Bannari Amman Institute of Technology Bannari Amman Institute of Technology Sathyamangalam, Erode Sathyamangalam, Erode [email protected] [email protected] Abstract—The DNA molecule is indubitably the most economic limits. And also that has produced small, faster powerful medium known for DNA’s ability to code, store chips, beyond an certain limit, the number of silicon atoms in information as a means of data storage. But till now, DNA the insulating layer of a transistor is no longer sufficient to molecule has found little use in computing applications. For prevent the leakage of electron that causes the circuit to shorten initiating computing application with DNA molecule, it requires [1]. For overcome these limitations the scientists and to design DNA transistors which can be utilized to design basic technologists are looking for new materials, innovative gates to implement Boolean logic. Interestingly some recent structures and revolutionary ideas to realize reliable transistor researches have shown that it’s very much possible to design a like action in such tiny space [2].Most novel materials three terminal transistor like device architecture by controlling available today are at the first step, where researchers are the flow of RNA polymerase along DNA with specific integrases. trying to understand their properties and characteristics of Along with that, very recently, fundamental experimental designs for realizing various basic Boolean logic functions have been transistors fabricated using them. -

From Genetic Engineering to Synthetic Biology
What & why is synthetic biology? U.S. Department of State Drew Endy Stanford Bioengineering The BioBricks Foundation 16 August 2013 Washington DC 1 Learn via building The first 70 years of molecular biology focused on taking living systems apart. We can also learn alot by putting molecular systems back together and testing how they behave relative to our expectations. [Note, policies and practices supporting scientific exchange and immigration were and remain key in making this happen.] 2 Make living matter fully programmable We also got really good at engineering information processing systems over the last 70 years. What should we aspire to master over the next 70 years? Engineerable living matter appears at the top of the list for many technologists. 3 As a fantastic example, the lawns and gardens of Menlo Park, CA grow 16 million pounds of state of the art nanotechnology each year. We mostly compost these materials. Could I reprogram a pinecone to grow computer chips instead? Photo by Roger Lancaster (http://www.flickr.com/photos/rogeral/5813079061/); educational fair use 44 Reprogramming pine trees will likely take too long. Perhaps instead partner with a wood fungus to manufacture materials from ground up yard waste (e.g., wood chips) Photo by Drew Endy during visit to Far West Fungi, Inc. near Monterey, CA with Phil Ross (philross.org) 55 20,000nm Cross section of biosilica spicule composite beam Add in biomineralization systems to assemble non- living objects via biomanufacturing platforms? Joanna Aizenberg et al., Science v.309 no.5732 pp.275-278, 8 July 2005 6 5,000,000nm (5mm) Biosilica cage Joanna Aizenberg et al., Science v.309 no.5732 pp.275-278, 8 July 2005 7 Develop new programming languages for controlling systems in space and time (i.e., programmed pattern formation). -

Engineering Bacterial Populations Via Dna Messaging
ENGINEERING BACTERIAL POPULATIONS VIA DNA MESSAGING ADISSERTATION SUBMITTED TO THE DEPARTMENT OF BIOENGINEERING AND THE COMMITTEE ON GRADUATE STUDIES OF STANFORD UNIVERSITY IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Monica Elise Ortiz June 2013 © 2013 by Monica Elise Ortiz. All Rights Reserved. Re-distributed by Stanford University under license with the author. This work is licensed under a Creative Commons Attribution- Noncommercial 3.0 United States License. http://creativecommons.org/licenses/by-nc/3.0/us/ This dissertation is online at: http://purl.stanford.edu/ht577gs4955 ii I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Andrew Endy, Primary Adviser I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Daniel Fisher I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Hans Riedel-Kruse Approved for the Stanford University Committee on Graduate Studies. Patricia J. Gumport, Vice Provost Graduate Education This signature page was generated electronically upon submission of this dissertation in electronic format. An original signed hard copy of the signature page is on file in University Archives. iii iv Abstract Evolution has selected for organisms that benefit from genetically encoded cell-cell communication. We observe cell-cell communication throughout every scale in na- ture, from simple single-celled bacteria to complex multicellular mammals. -

Wetware Technology (Technology of the Future)
International Journal of Recent Advances in Engineering & Technology (IJRAET) _______________________________________________________________________________________________ WETWARE TECHNOLOGY (TECHNOLOGY OF THE FUTURE) 1V.Nivas Prabu, 2J. Gowtham Kumar 1,2CSE (department), Panimalar Institute of Technology, Chennai, India. Email: [email protected], [email protected] Abstract: New generation of fast and flexible computers continuously changing physical connections, and that can work out for themselves how to solve a problem, chemical and electrical influences that spread across the rather than having to be told exactly what to do. Ordinary body[Fig:1.1]. The process by which the mind and brain computers need absolutely correct information every time interact to produce the collection of experiences that we to come to the right answer. A biological computer will define as self-awareness is still seriously in question. come to the correct answer based on partial information, by filling in the gaps itself. The device the team has built A cell’s survival depends on how well it can detect and can "think for itself" because the neurons are able to form respond to environmental cues. In this sense, a cell is an their own connections from one to another. Normal silicon organically derived computer — it takes input from its computers only make the connections they are told to by surroundings and uses logic circuits to result in a the programmer. This flexibility means the biological computer works out it own way of solving the problem. specific output. The central argument of Wetwareis that an individual cell contains thousands of enzymes, each I. INTRODUCTION TO WETWARE: performing reiterative, molecular processes. -

Molecular Computing: Paths to Chemical Turing Machines Cite This: Chem
Chemical Science View Article Online PERSPECTIVE View Journal | View Issue Molecular computing: paths to chemical Turing machines Cite this: Chem. Sci.,2015,6,6050 Shaji Varghese,* Johannes A. A. W. Elemans, Alan E. Rowan and Roeland J. M. Nolte* To comply with the rapidly increasing demand of information storage and processing, new strategies for computing are needed. The idea of molecular computing, where basic computations occur through molecular, supramolecular, or biomolecular approaches, rather than electronically, has long captivated researchers. The prospects of using molecules and (bio)macromolecules for computing is not without precedent. Nature is replete with examples where the handling and storing of data occurs with high efficiencies, low energy costs, and high-density information encoding. The design and assembly of computers that function according to the universal approaches of computing, such as those in a Turing machine, might be realized in a chemical way in the future; this is both fascinating and extremely Received 26th June 2015 challenging. In this perspective, we highlight molecular and (bio)macromolecular systems that have been Accepted 6th August 2015 designed and synthesized so far with the objective of using them for computing purposes. We also Creative Commons Attribution 3.0 Unported Licence. DOI: 10.1039/c5sc02317c present a blueprint of a molecular Turing machine, which is based on a catalytic device that glides along www.rsc.org/chemicalscience a polymer tape and, while moving, prints binary information