Nectarinia: Nectariniidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MADAGASCAR: the Wonders of the “8Th Continent” a Tropical Birding Custom Trip
MADAGASCAR: The Wonders of the “8th Continent” A Tropical Birding Custom Trip October 20—November 6, 2016 Guide: Ken Behrens All photos taken during this trip by Ken Behrens Annotated bird list by Jerry Connolly TOUR SUMMARY Madagascar has long been a core destination for Tropical Birding, and with the opening of a satellite office in the country several years ago, we further solidified our expertise in the “Eighth Continent.” This custom trip followed an itinerary similar to that of our main set-departure tour. Although this trip had a definite bird bias, it was really a general natural history tour. We took our time in observing and photographing whatever we could find, from lemurs to chameleons to bizarre invertebrates. Madagascar is rich in wonderful birds, and we enjoyed these to the fullest. But its mammals, reptiles, amphibians, and insects are just as wondrous and accessible, and a trip that ignored them would be sorely missing out. We also took time to enjoy the cultural riches of Madagascar, the small villages full of smiling children, the zebu carts which seem straight out of the Middle Ages, and the ingeniously engineered rice paddies. If you want to come to Madagascar and see it all… come with Tropical Birding! Madagascar is well known to pose some logistical challenges, especially in the form of the national airline Air Madagascar, but we enjoyed perfectly smooth sailing on this tour. We stayed in the most comfortable hotels available at each stop on the itinerary, including some that have just recently opened, and savored some remarkably good food, which many people rank as the best Madagascar Custom Tour October 20-November 6, 2016 they have ever had on any birding tour. -

Biolphilately Vol-64 No-3
BIOPHILATELY OFFICIAL JOURNAL OF THE BIOLOGY UNIT OF ATA MARCH 2020 VOLUME 69, NUMBER 1 Great fleas have little fleas upon their backs to bite 'em, And little fleas have lesser fleas, and so ad infinitum. —Augustus De Morgan Dr. Indraneil Das Pangolins on Stamps More Inside >> IN THIS ISSUE NEW ISSUES: ARTICLES & ILLUSTRATIONS: From the Editor’s Desk ......................... 1 Botany – Christopher E. Dahle ............ 17 Pangolins on Stamps of the President’s Message .............................. 2 Fungi – Paul A. Mistretta .................... 28 World – Dr. Indraneil Das ..................7 Secretary -Treasurer’s Corner ................ 3 Mammalia – Michael Prince ................ 31 Squeaky Curtain – Frank Jacobs .......... 15 New Members ....................................... 3 Ornithology – Glenn G. Mertz ............. 35 New Plants in the Philatelic News of Note ......................................... 3 Ichthyology – J. Dale Shively .............. 57 Herbarium – Christopher Dahle ....... 23 Women’s Suffrage – Dawn Hamman .... 4 Entomology – Donald Wright, Jr. ........ 59 Rats! ..................................................... 34 Event Calendar ...................................... 6 Paleontology – Michael Kogan ........... 65 New Birds in the Philatelic Wedding Set ........................................ 16 Aviary – Charles E. Braun ............... 51 Glossary ............................................... 72 Biology Reference Websites ................ 69 ii Biophilately March 2020 Vol. 69 (1) BIOPHILATELY BIOLOGY UNIT -

Nature Protection Trust of Seychelles
NATURE PROTECTION TRUST OF SEYCHELLES RESEARCH AND MONITORING ANNUAL REPORT 2010 TABLE OF CONTENTS ACKNOWLEDGEMENTS 1 A. ORGANISATION BACKGROUND AND CAPACITY 2 B. RESEARCH AND MONITORING 2 1. SUMMARY 2 2. SPECIES 2 2.1. Birds 2 2.2. Reptiles 3 2.3. Land and Marine Mammals 7 2.4. Amphibians 8 2.5. Invertebrates 10 2.6. Fish and other freshwater/marine species 17 2.7. Plants 20 3. HABITATS 21 3.1. Marine 21 3.2. Forests 23 3.3. Wetland 26 3.4. Climate 26 4. ENVIRONMENTAL MANAGEMENT 28 ACKNOWLEDGEMENTS NPTS is grateful to Labriz in logistical support and assistance with forest restoration. We are also grateful to the Sussex University School of Geography for monitoring assistance. 1 A. ORGANISATION BACKGROUND AND CAPACITY Nature Protection Trust of Seychelles was established in 1992 as a non-profit-making, non-governmental organisation dedicated to furthering the conservation of nature in the Seychelles islands, through informed scientific research to conserve species by conserving their habitats. Fulfilling its aim of promoting education, study and research in all fields of natural history the NPTS publishes two journals. The annual scientific journal, “Phelsuma” deals with all aspects of nature within the entire western Indian Ocean and includes reports on all NPTS projects. “Birdwatch” is a quarterly publication containing news and anecdotes concerning the natural history in the islands. NPTS manages the Silhouette Conservation Project (ecosystem conservation and research) and the Seychelles Giant Tortoise and Seychelles Terrapin Conservation Projects (research, captive breeding and reintroduction). Within the Silhouette Conservation Project there are special research projects on Seychelles sheath-tailed bats, sooglossid frogs and climate change. -

MADAGASCAR: the Wonders of the “8Th Continent” a Tropical Birding Set Departure
MADAGASCAR: The Wonders of the “8th Continent” A Tropical Birding Set Departure November 3—28, 2013 Guide: Ken Behrens All photos taken during this trip. All photos by Ken Behrens unless noted otherwise. TOUR SUMMARY Madagascar has long been a core destination for Tropical Birding, and with last year’s opening of a satellite office in the country, we have further solidified our expertise in the “Eighth Continent.” This was another highly successful set-departure tour to this special island. It included both the Northwestern Endemics Pre-Trip at the start and the Helmet Vanga extension to the Masoala Peninsula at the end. Although Madagascar poses some logistical challenges, especially in the form of the national airline Air Madagascar, we had no problems on this tour, not even a single delayed flight! The birding was great, with 196 species recorded, including almost all of the island’s endemic birds. As usual, the highlight was seeing all five of the incredible ground-rollers, from the roadrunner-like Long-tailed of the spiny forest to the wonderful rainforest-dwelling Scaly. There was a strong cast of vangas, including Helmet, Bernier’s, and Sickle-billed. In fact, we saw every member of the family save the mysterious Red-tailed Newtonia which is only regularly seen in the far south. As normal, the couas were also a favorite. From the shy and beautiful Red-breasted of Madagascar Set Departure Tour Nov. 3-28, 2013 the eastern rainforest to the huge Giant Coua of the dry western forest, we were looking for and at couas virtually every day! The bizarre mesites form a Malagasy endemic family, and we had superb extended views of all three members of the family. -

Atoll Research Bulletin No. 252 Bird and Denis Islands
ATOLL RESEARCH BULLETIN NO. 252 BIRD AND DENIS ISLANDS, SEYCHELLES by D. R. Stoddart and F. R. Fosberg Issued by THE SMITHSONIAN INSTITUTION Washington, D. C., U.S.A. ~ul~'l981 Contents 1. Geography and ecology of Bird Island, Seychelles Introduction Morphology and structure Climate Vegetation Flora Invertebrates Reptiles Mammals Birds History 2. Plants recorded from Bird Island 3. Geography and ecology of Denis Island, Seychelles Introduction Morphology and structure Climate Vegetation Flora Invertebrates Reptiles Mammals Birds History 4. Plants recorded from Denis Island 5. References Manuscript received May 1980 --Eds. List of Figures 1. The Seychelles Bank following page 11 2. Bird Island in 1976 following page 11 3. Beach sediment at Bird Island following page 11 4. Denis Island in 1977 following page 50 5. Monthly rainfall at Denis Island, 19 71-1962 following page 50 List of Tables 1. Scientific studies at Bird Island 2. Characteristics of Bird Island beach sands 3. Monthly rainfall at Bird Island, 1951-1962 4. Key to the literature on insects collected at Bird Island 5. Scientific studies at Denis Island 6. Monthly and annual rainfall records at Denis Island iii List of Plates Bird Island: Suriana zone on the northeast shore following page 11 Bird Island: Pisonia and Cordia woodland with Suriana on the northeast shore Bird Island: Tournefortia parkland in the northeast Bird Island: tree-like Tournefortia in the northeast Bird Island: pioneer sedges and Scaevola on the east shore Bird Island: pioneer Ipomoea pes-caprae on the east shore Bird Island: pioneer sedges, Scaevola and Tournefortia on the northeast shore Bird Island: airstrip from the southeast Denis Island: phosphate cliffs with Casuarina woodland, southwest shore following page 50 10. -

Madagascar Highlights Knox: Private 28Th October to 11Th November 2016 (15 Days) Trip Report
Madagascar Highlights Knox: Private 28th October to 11th November 2016 (15 days) Trip Report Short-legged Ground Roller by Gareth Robbins Trip report and photos by tour leader: Gareth Robbins Trip Report - Knox Private Madagascar Highlights 2016 2 ________________________________________________________________________________ Tour Summary Mike and Linn arrived on a prompt flight in from South Africa and once they made it through customs, they were greeted by Guillaume - who is the MEA (Madagascan Expedition Agency a Representative and Guide) and myself. We made a few stops along the way to the hotel and had a brief glimpse of Malagasy Kestrel as it shot over the road. We finally arrived at our beautiful, cool hotel called Au Bois Vert, which lay at the foot of a cluster of pine trees. There were a couple of birds spotted on the property after check-in, such as Malagasy Bulbul, Red Fody, Malagasy Brush Warbler, Madagascan Wagtail and Malagasy White-eye. After a lovely dinner, we made our way back to our rooms and were serenaded by the Madagascan Wagtail by Gareth Robbins Madagascan Nightjar throughout the night. We said goodbye to our peaceful and serene hotel and made our way through the hustle and bustle of the city of 1000 soldiers - Antananarivo. After leaving the city, we drove through a few smaller villages, managing to spot Dimorphic and Western Cattle Egrets, and Squacco and Striated Herons, as well as a pair of Malagasy Kingfishers in some of the rice paddies we passed. We finally arrived at our hotel along the river in Perinet and as we walked to reception, we spotted a Malagasy Paradise Flycatcher in the trees in front of the restaurant. -

Seychelles & Madagascar with Aldabra Atoll
SEYCHELLES & MADAGASCAR WITH ALDABRA ATOLL ABOARD THE SILVER DISCOVERER OCTOBER 20–NOVEMBER 6, 2018 Pitta-like Ground Roller was found near a freshly dug nest in Amber Mountain NP (Photo M. Valkenburg) LEADER: MACHIEL VALKENBURG LIST COMPILED BY: MACHIEL VALKENBURG VICTOR EMANUEL NATURE TOURS, INC. 2525 WALLINGWOOD DRIVE, SUITE 1003 AUSTIN, TEXAS 78746 WWW.VENTBIRD.COM By Machiel Valkenburg When I learned about the option of leading this fascinating Zegrahm cruise for VENT, I jumped right on it, as I knew it would deliver some spectacular birds, mammals, and landscapes. Oooh man I was right! The trip started in paradisiacal surroundings on the island of Mahe, the largest of all islands that together form the Seychelles. Before embarking the Silver Discoverer , we planned some pre-tour birding. In the afternoon we made an outing to find the rare Seychelles White-eye, Seychelles Blue-Pigeon, and Seychelles Sunbird. Rapidly they were found, and good looks were enjoyed by all. In the evening after dinner we went out for probably the rarest and most difficult bird of the entire tour, the Seychelles Scops-Owl. For a long time we heard no response, but just before we decided to leave the designated spot we heard one quick rasping call. Soon after, we found the bird in full delight—just amazing! The enchanting Seychelles Sunbird has a very restricted range (photo M.Valkenburg) Our journey would take us from several Seychellois islands to Madagascar, visiting the world-famous Aldabra Atoll on the way, ending the trip with visits to Reunion and Mauritius. This adventure was one-of- a-kind, with our program visiting so many difficult to reach places where we had the chance of finding many of the world’s most hard-to-get endangered species. -

Birds Calling but Couldn’T Lure Any Closer During the Heat of the Mid Afternoon
Madagascar - Tom and Linda Arny 7th to 18th November 2007 Trip Report compiled by James Currie Tour Summary The tour started on the 7th November after meeting Tom and Linda at Johannesburg International Airport. We arrived in Tana in the late afternoon and commenced the journey to Perinet, ticking our first selection of egrets and herons in the Tana wetlands. It was dark by the time we arrived at Perinet and we embarked on a short evening walk, hearing both Long-eared Owl and Rainforest Scops Owl. The next morning saw us visiting Perinet Special Reserve where we were treated to superb views of Indri. We spent 3 nights in the Perinet/Mantadia area and visited Mantadia on two consecutive mornings, with the balance of the time being spent at Perinet. Highlights from Mantadia included crippling views of Madagascar Pygmy Kingfisher, nesting Velvet Asity and Pitta-like and Scaly Ground-Rollers with particularly good looks at the latter species, several good views of Nuthatch Vanga climbing up and down tree trunks, a pair of Madagascar Little Grebes, stunning looks at Greater Vasa Parrot, Madagascar Blue Pigeon, a greenbul flock that included Grey-crowned, Longbilled and Spectacled Greenbuls with a brief sighting of an unconfirmed Madagascar Yellowbrow, a troop of confiding Diademed Sifakas with Rufous-headed Ground-Roller calling in the background. Highlights from RBT Madagascar Trip Report 2007 2 Perinet included superb close-up views of both Red-breasted Coua and Madagascar Wood Rail, roosting Collared Nightjar, Madagascar Long-eared Owl, skulking Madagascar Flufftail, Red-fronted Coua, nesting Chabert’s and White-headed Vangas and a roosting Rainforest Scops Owl. -

Miscellaneous Behavioural Observations of Malagasy Birds
MADAGASCAR CONSERVATION & DEVELOPMENT VOLUME 1 3 | ISSUE 01 — DECEMBER 201 8 PAGE 70 SHORT NOTE http://dx.doi.org/1 0.431 4/mcd.v1 3i1 .8 Miscellaneous behavioural observations of Malagasy birds Charlie J. GardnerI and Louise D. Jasper Correspondence: Charlie J. Gardner Durrell Institute of Conservation and Ecology (DICE) School of Anthropology and Conservation, University of Kent United Kingdom Email: [email protected] ABSTRACT piégeage d’un Capucin de Madagascar Lepidopygia nana dans Madagascar possesses a unique avifauna characterized by high une toile d’araignée (Nephila sp.), et vii) le harcèlement des rep- endemism rates at species and higher taxonomic levels, but little tiles incluant des serpents (Ithycyphus miniatus, Acrantophis is known about the behaviour, diets and interspecific interactions madagascariensis) et un caméléon (Furcifer pardalis) par le of many species. We present a number of opportunistic observa- Souimanga malgache Nectarinia souimanga, le Shama de tions of Malagasy birds collected during 201 2–201 5, including a Madagascar Copsychus albospecularis, le Tchitrec malgache foraging association between Hook-billed vanga Vanga curvirostris Terpsiphone mutata, la Newtonie commune Newtonia and White-breasted mesite Mesitornis variegatus, aggressive in- brunneicauda et le Drongo malgache Dicrurus forficatus. teraction between a fledgling Madagascar cuckoo Cuculus rochii and its Common jery Neomixis tenella host, records of carnivory in Green-capped coua Coua ruficeps olivaceiceps and frugivory in Lafresnaye’s vanga Xenopirostris xenopirostris, an unusual aggre- gation of Alpine swift Tachymarptis melba around a telecommuni- Madagascar possesses a unique avifauna characterized by high cations tower, entrapment of Madagascar mannikin Lepidopygia levels of endemism at species and higher taxonomic levels. -

THE BEST of MADAGASCAR and NORTHWEST and BERENTY EXTENSIONS TRIP REPORT 15 September ‐ 5 October 2016 by Justin Nicolau
THE BEST OF MADAGASCAR AND NORTHWEST AND BERENTY EXTENSIONS TRIP REPORT 15 September ‐ 5 October 2016 By Justin Nicolau Giant Coua - Coua gigas www.birdingecotours.com [email protected] [email protected] 2 | T R I P R E P O R T MADAGASCAR, 2016 ITINERARY Day Date Start Finish 1 15 September 2016 Antananarivo Antananarivo 2 16 September 2016 Antananarivo Antananarivo 3 17 September 2016 Antananarivo Mahajanga 4 18 September 2016 Mahajanga Ankarafantsika NP 5 19 September 2016 Ankarafantsika NP Ankarafantsika NP 6 20 September 2016 Ankarafantsika NP Mahajanga 7 21 September 2016 Mahajanga Antananarivo 8 22 September 2016 Antananarivo Berenty Reserve 9 23 September 2016 Berenty Reserve Berenty Reserve 10 24 September 2016 Berenty Reserve Ifaty 11 25 September 2016 Ifaty Ifaty 12 26 September 2016 Ifaty Toliara 13 27 September 2016 Toliara Isalo 14 28 September 2016 Isalo Ranomafana NP 15 29 September 2016 Ranomafana NP Ranomafana NP 16 30 September 2016 Ranomafana NP Antsirabe 17 01 October 2016 Antsirabe Andasibe 18 02 October 2016 Andasibe Andasibe 19 03 October 2016 Andasibe Andasibe 20 04 October 2016 Andasibe Antananarivo 21 05 October 2016 Antananarivo Departure Day 1: 15 September 2016 Arriving in Antananarivo mid afternoon, the group set out after the meets and greets and currency exchange toward our hotel for the next two nights. En route the bird sightings were few and far between, as one would expect while in transit. We picked up Common Myna, Dimorphic Egret, Squacco Heron, and a few Mascarene Martins. Malagasy Kingfisher - Corythornis vintsioides www.birdingecotours.com [email protected] [email protected] 3 | T R I P R E P O R T MADAGASCAR, 2016 Arriving at the hotel, bags offloaded, and room keys in hand, we set off for a brief walk around a small body of water in front of the accommodation. -
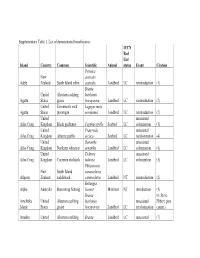
Supplementary Table 1. List of Demonstrated Beneficiaries
Supplementary Table 1. List of demonstrated beneficiaries. IUCN Red List Island Country Common Scientific Animal status Event Citation Petroica New australis Adele Zealand South Island robin australis Landbird LC reintroduction (1) Branta United Aleutian cackling hutchinsii Agattu States goose leucopareia Landbird LC reintroduction (2) United Evermann's rock Lagopus muta Agattu States ptarmigan evermanni Landbird LC reintroduction (2) United unassisted Ailsa Craig Kingdom Black guillemot Cepphus grylle Seabird LC colonization (3) United Fratercula unassisted Ailsa Craig Kingdom Atlantic puffin arctica Seabird LC recolonization (4) United Oenanthe unassisted Ailsa Craig Kingdom Northern wheatear oenanthe Landbird LC colonization (4) United Tadorna unassisted Ailsa Craig Kingdom Common shelduck tadorna Landbird LC colonization (3) Philesturnus New South Island carunculatus Allports Zealand saddleback carunculatus Landbird NT reintroduction (2) Bettongia Alpha Australia Burrowing bettong lesueur Mammal NT introduction (5) Branta (6; Steve Amchitka United Aleutian cackling hutchinsii unassisted Ebbert, pers. Island States goose leucopareia Landbird LC recolonization comm.) Amukta United Aleutian cackling Branta Landbird LC unassisted (7) IUCN Red List Island Country Common Scientific Animal status Event Citation States goose hutchinsii recolonization leucopareia Sally Amy Poncet, Island/Outer United Cinclodes unassisted unpublished Knob Kingdom Tussacbird antarcticus Landbird LC recolonization data Sally Amy Poncet, Island/Outer United unpublished -

Madagascar Budget Birding 10Th October to 20Th October 2021 (11 Days)
Madagascar Budget Birding 10th October to 20th October 2021 (11 days) Pitta-like Ground Roller by Markus Lilje Our Budget Madagascar Birding tour provides the budget conscious birder with a fantastic opportunity for observing the highlights of this magical island’s incredible birds and wildlife. On this tour we will visit three of Madagascar's core critical habitats; namely the deciduous woodlands of Ankarafantsika, the dry spiny forest around Ifaty and the moist eastern rainforests of Analamazaotra and Mantadia (Perinet), all of which hold an exceptional diversity of endemic birds and lemurs. We stand excellent chances of finding all 5 of the island's endemic bird families, as well as up to 10 different lemur species, including the awesome Indri! Some of the expected highlights of this tour include Long-tailed and Pitta- like Ground Rollers, stunning Schlegel's and Velvet Asities, Blue Coua, Subdesert and White-breasted Mesites, Sickle-billed and Van Dam's Vangas, and the ubiquitous Cuckoo Roller. RBL Madagascar – Budget Birding Itinerary 2 THE TOUR AT A GLANCE… THE ITINERARY Day 1 Antananarivo and drive to Perinet Days 2 & 3 Perinet Special Reserve and Mantadia National Park Day 4 Perinet drive to Antananarivo Day 5 Antananarivo to Mahajanga and transfer to Ampijoroa Forest Station Day 6 Ampijoroa Forest Station Day 7 Ampijoroa to Mahajanga and flight to Antananarivo Day 8 Antananarivo to Ifaty via Tulear Day 9 Ifaty Day 10 Ifaty to Tulear Day 11 Tulear to Antananarivo and depart TOUR ROUTE MAP… RBL Madagascar – Budget Birding Itinerary 3 THE TOUR IN DETAIL… Day 1: Antananarivo and drive to Perinet.