Impaired T Lymphocyte Function Increases Tumorigenicity and Decreases Tumor Latency in a Mouse Model of Head and Neck Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hepatokines As a Molecular Transducer of Exercise
Journal of Clinical Medicine Review Hepatokines as a Molecular Transducer of Exercise Dae Yun Seo 1 , Se Hwan Park 2 , Jubert Marquez 1, Hyo-Bum Kwak 3 , Tae Nyun Kim 1 , Jun Hyun Bae 4 , Jin-Ho Koh 5 and Jin Han 1,* 1 National Research Laboratory for Mitochondrial Signaling, Department of Physiology, BK21 Plus Project Team, College of Medicine, Smart Marine Therapeutics Center, Cardiovascular and Metabolic Disease Center, Inje University, Busan 47392, Korea; [email protected] (D.Y.S.); [email protected] (J.M.); [email protected] (T.N.K.) 2 School of Kinesiology, Yeungnam University, Gyeongsan 38541, Korea; [email protected] 3 Department of Kinesiology, Inha University, Incheon 22212, Korea; [email protected] 4 Institute of Sport Science, Seoul National University, Seoul 08826, Korea; [email protected] 5 Department of Physiology, College of Medicine, Yeungnam University, Daegu 42415, Korea; [email protected] * Correspondence: [email protected]; Tel.: +82-051-890-6727 Abstract: Exercise has health benefits and prevents a range of chronic diseases caused by physiologi- cal and biological changes in the whole body. Generally, the metabolic regulation of skeletal muscle through exercise is known to have a protective effect on the pathogenesis of metabolic syndrome, non-alcoholic fatty liver disease (NAFLD), type 2 diabetes (T2D), and cardiovascular disease (CVD). Besides this, the importance of the liver as an endocrine organ is a hot research topic. Hepatocytes also secrete many hepatokines in response to nutritional conditions and/or physical activity. In particular, certain hepatokines play a major role in the regulation of whole-body metabolic homeosta- sis. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Zinc-Binding to Leukocyte Cell Derived Chemotaxin 2 and Implications for Its Associated Amyloidosis by Jethro Emmanuel Prinston
Zinc-binding to leukocyte cell derived chemotaxin 2 and implications for its associated amyloidosis By Jethro Emmanuel Prinston A thesis submitted in conformity with the requirements for the degree of Master of Science Graduate Department of Biochemistry University of Toronto ©Copyright Jethro Emmanuel Prinston, 2020 i Zinc-binding to leukocyte cell derived chemotaxin 2 and implications for its associated amyloidosis Jethro Emmanuel Prinston Master of Science Graduate Department of Biochemistry University of Toronto 2020 Abstract Deposition of leukocyte cell-derived chemotaxin 2 (LECT2) as amyloid (ALECT2) leads to ALECT2 amyloidosis, an amyloid disease with no known cure. Recent findings indicate that ALECT2 amyloidosis is severely underdiagnosed. All sequenced ALECT2 amyloidosis patients have a single nucleotide polymorphism that changes an isoleucine (WT) to a valine (58V) at position 58. Currently, little is known of the mechanism by which the protein misfolds and aggregates. Herein, we demonstrate the first molecular characterization of the misfolding and aggregation of the mature LECT2 protein. Through various biophysical methods, we found that zinc-binding alters the secondary structure, increases the conformational stability, and reduces aggregation propensity of both WT and 58V-LECT2. No significant difference was found between the variants in their secondary structure, conformational stability, and zinc-binding affinity. However, zinc-free 58V-LECT2 was significantly more amyloidogenic than WT-LECT2. Therapies aimed at ALECT2 amyloidosis should exploit putative conformational differences between zinc- bound and zinc-free LECT2. ii Acknowledgements This study would not be made possible without the support of several individuals. I thank current and previous members of the Chakrabartty Lab: Dr Natalie Galant, Meghan Wing, Dr. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Downloaded in Aug 2019
ARTICLE https://doi.org/10.1038/s41467-021-24473-2 OPEN IL-15 and PIM kinases direct the metabolic programming of intestinal intraepithelial lymphocytes Olivia J. James1, Maud Vandereyken1, Julia M. Marchingo 2, Francois Singh1, Susan E. Bray3, Jamie Wilson4, ✉ Andrew G. Love1 & Mahima Swamy 1 1234567890():,; Intestinal intraepithelial lymphocytes (IEL) are an abundant population of tissue-resident T cells that protect and maintain the intestinal barrier. IEL respond to epithelial cell-derived IL-15, which is complexed to the IL-15 receptor α chain (IL-15/Rα). IL-15 is essential both for maintaining IEL homeostasis and inducing IEL responses to epithelial stress, which has been associated with Coeliac disease. Here, we apply quantitative mass spectrometry to IL-15/Rα- stimulated IEL to investigate how IL-15 directly regulates inflammatory functions of IEL. IL-15/ Rα drives IEL activation through cell cycle regulation, upregulation of metabolic machinery and expression of a select repertoire of cell surface receptors. IL-15/Rα selectively upregu- lates the Ser/Thr kinases PIM1 and PIM2, which are essential for IEL to proliferate, grow and upregulate granzyme B in response to inflammatory IL-15. Notably, IEL from patients with Coeliac disease have high PIM expression. Together, these data indicate PIM kinases as important effectors of IEL responses to inflammatory IL-15. 1 MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee, Dundee, UK. 2 Division of Cell Signalling and Immunology, School of Life Sciences, University of Dundee, Dundee, UK. 3 NHS Research Scotland, Tayside Tissue Biorepository, University of Dundee, Dundee, UK. 4 Department of ✉ Pathology, NHS Tayside, Ninewells Hospital, Dundee, UK. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Supplementary Table S5. Differentially Expressed Gene Lists of PD-1High CD39+ CD8 Tils According to 4-1BB Expression Compared to PD-1+ CD39- CD8 Tils
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Supplementary Table S5. Differentially expressed gene lists of PD-1high CD39+ CD8 TILs according to 4-1BB expression compared to PD-1+ CD39- CD8 TILs Up- or down- regulated genes in Up- or down- regulated genes Up- or down- regulated genes only PD-1high CD39+ CD8 TILs only in 4-1BBneg PD-1high CD39+ in 4-1BBpos PD-1high CD39+ CD8 compared to PD-1+ CD39- CD8 CD8 TILs compared to PD-1+ TILs compared to PD-1+ CD39- TILs CD39- CD8 TILs CD8 TILs IL7R KLRG1 TNFSF4 ENTPD1 DHRS3 LEF1 ITGA5 MKI67 PZP KLF3 RYR2 SIK1B ANK3 LYST PPP1R3B ETV1 ADAM28 H2AC13 CCR7 GFOD1 RASGRP2 ITGAX MAST4 RAD51AP1 MYO1E CLCF1 NEBL S1PR5 VCL MPP7 MS4A6A PHLDB1 GFPT2 TNF RPL3 SPRY4 VCAM1 B4GALT5 TIPARP TNS3 PDCD1 POLQ AKAP5 IL6ST LY9 PLXND1 PLEKHA1 NEU1 DGKH SPRY2 PLEKHG3 IKZF4 MTX3 PARK7 ATP8B4 SYT11 PTGER4 SORL1 RAB11FIP5 BRCA1 MAP4K3 NCR1 CCR4 S1PR1 PDE8A IFIT2 EPHA4 ARHGEF12 PAICS PELI2 LAT2 GPRASP1 TTN RPLP0 IL4I1 AUTS2 RPS3 CDCA3 NHS LONRF2 CDC42EP3 SLCO3A1 RRM2 ADAMTSL4 INPP5F ARHGAP31 ESCO2 ADRB2 CSF1 WDHD1 GOLIM4 CDK5RAP1 CD69 GLUL HJURP SHC4 GNLY TTC9 HELLS DPP4 IL23A PITPNC1 TOX ARHGEF9 EXO1 SLC4A4 CKAP4 CARMIL3 NHSL2 DZIP3 GINS1 FUT8 UBASH3B CDCA5 PDE7B SOGA1 CDC45 NR3C2 TRIB1 KIF14 TRAF5 LIMS1 PPP1R2C TNFRSF9 KLRC2 POLA1 CD80 ATP10D CDCA8 SETD7 IER2 PATL2 CCDC141 CD84 HSPA6 CYB561 MPHOSPH9 CLSPN KLRC1 PTMS SCML4 ZBTB10 CCL3 CA5B PIP5K1B WNT9A CCNH GEM IL18RAP GGH SARDH B3GNT7 C13orf46 SBF2 IKZF3 ZMAT1 TCF7 NECTIN1 H3C7 FOS PAG1 HECA SLC4A10 SLC35G2 PER1 P2RY1 NFKBIA WDR76 PLAUR KDM1A H1-5 TSHZ2 FAM102B HMMR GPR132 CCRL2 PARP8 A2M ST8SIA1 NUF2 IL5RA RBPMS UBE2T USP53 EEF1A1 PLAC8 LGR6 TMEM123 NEK2 SNAP47 PTGIS SH2B3 P2RY8 S100PBP PLEKHA7 CLNK CRIM1 MGAT5 YBX3 TP53INP1 DTL CFH FEZ1 MYB FRMD4B TSPAN5 STIL ITGA2 GOLGA6L10 MYBL2 AHI1 CAND2 GZMB RBPJ PELI1 HSPA1B KCNK5 GOLGA6L9 TICRR TPRG1 UBE2C AURKA Leem G, et al. -

Circulating LECT2 Levels in Newly Diagnosed Type 2 Diabetes Mellitus
® Observational Study Medicine OPEN Circulating LECT2 levels in newly diagnosed type 2 diabetes mellitus and their association with metabolic parameters An observational study Zhen Zhang, PhDa, Huixian Zeng, MDb, Jianghong Lin, MDc, Yinghui Hu, MDd, Rui Yang, PhDa, ∗ ∗ Jia Sun, PhDa, Rongping Chen, PhDa, , Hong Chen, PhDa, Abstract Leukocyte cell-derived chemotaxin 2 (LECT2) is a hepatokine expressed in hepatocytes and appears to be involved in energy metabolism. The aim of this study was to determine plasma LECT2 levels in newly diagnosed type 2 diabetic patients and to correlate the results with various metabolic parameters. A total of 93 newly diagnosed type 2 diabetic patients and 80 age- and sex-matched nondiabetes mellitus ones were enrolled in the study. Plasma LECT2 levels were measured by enzyme-linked immunosorbent assay. Circulating LECT2 levels were approximately 1.3 times higher in newly diagnosed type 2 diabetic patients than in controls (mean 30.30 vs 23.23ng/mL, P < .001). Correlation analysis showed that LECT2 was negatively associated with high-density lipoprotein- cholesterol (HDL-C) levels in type 2 diabetic patients and obese subjects (P < .05). In multiple stepwise regression analysis, HDL-C, HOMA-IR, BMI, FINS, and TG were significantly independent determinants for LECT2 (P < .05). Our study showed that circulating LECT2 concentrations are significantly higher in newly diagnosed type 2 diabetic patients and further elevated in obese type 2 diabetic patients. LECT2 concentrations are significantly negatively associated -

32-13307: LY9 Human Description Product Info
9853 Pacific Heights Blvd. Suite D. San Diego, CA 92121, USA Tel: 858-263-4982 Email: [email protected] 32-13307: LY9 Human Lymphocyte Antigen 9, Signaling Lymphocytic Activation Molecule 3, Cell Surface Molecule Ly-9, SLAM Alternative Name : Family Member 3, SLAMF3, CD229 Antigen, CD229, Hly9, MLY9, LY9. Description Source: Sf9, Baculovirus cells. Sterile filtered colorless solution. Lymphocyte Antigen 9, also known as LY9 is a member of the SLAM family of immunomodulatory receptors, LY9 interacts with the adaptor molecule SAP . LY9 plays a part in adhesion reactions between T lymphocytes and accessory cells via homophilic interaction. LY9 promotes T-cell differentiation into a helper T-cell Th17 phenotype which leads to increased IL-17 secretion; the costimulatory activity requires SH2D1A. Furthermore it also promotes recruitment of RORC to the IL-17 promoter. LY9 is implicated in the maintenance of peripheral cell tolerance through serving as a negative regulator of the immune response. LY9 disables autoantibody responses as well as inhibits IFN-gamma secretion by CD4(+) T-cells. Moreover, LY9 negatively regulate the size of thymic innate CD8(+) T-cells and the development of invariant natural killer T (iNKT) cells. Systemic Lupus Erythematosus is one of the diseases which are associated with LY9. LY9 Human Recombinant produced in Sf9 Baculovirus cells is a single, glycosylated polypeptide chain containing 416 amino acids (48-454 aa) and having a molecular mass of 45.9kDa (Migrates at 40-57kDa on SDS-PAGE under reducing conditions).LY9 is expressed with a 6 amino acid His tag at C-Terminus and purified by proprietary chromatographic techniques. -

Quantitative Proteomic Approaches in a Zebrafish Model of Granulocytic Inflammation
Quantitative Proteomic Approaches in a Zebrafish Model of Granulocytic Inflammation Zur Erlangung des akademischen Grades eines DOKTORS DER NATURWISSENSCHAFTEN (Dr. rer. nat.) Fakultät für Chemie und Biowissenschaften Karlsruher Institut für Technologie (KIT) – Universitätsbereich Genehmigte DISSERTATION Von Sachin Kumar Singh Indian 2015 Dekan: Prof. Dr. Peter Roesky Referent: Prof. Dr. Andrew Cato Korreferent: Prof. Dr. Martin Bastmeyer Tag der mündlichen Prüfung: 10.02.2015 I hereby declare that this dissertation is my own independent work. I have only used the given sources and materials. I have cited others work appropriately. Sachin Singh Karlsruhe 02.01.2015 Abstract Abstract Neutrophils represent the most abundant type of leukocytes in vertebrates and are the first cells migrating to sites of trauma. Despite their fundamental importance, our understanding of the molecular changes occurring within neutrophils upon inflammation in a living animal remains fairly limited. In this study, the zebrafish model was used to investigate the in vivo orchestration of a neutrophil inflammatory response at the protein level. Firstly, the proteome of resting whole kidney marrow (WKM) neutrophils was investigated. Resting WKM neutrophils express more than 1500 proteins. Of these, 75% are enriched in neutrophils compared to neutrophil free brain tissue. In addition, the zebrafish neutrophil proteome shows partial conservation of immune related proteins between zebrafish and human. Secondly, a chemically induced inflammation assay in adult zebrafish was established in order to investigate proteome changes within activated neutrophils. To this end, a highly accurate and precise quantitative proteomics approach was required allowing for relative quantification of protein levels between samples. Two proteomics approaches, the label free and the SILAC (stable isotope labeling by amino acids in cell culture) method were thus applied to investigate the proteome dynamics within neutrophils upon inflammation. -

Characteristics of B Cell-Associated Gene Expression in Patients With
MOLECULAR MEDICINE REPORTS 13: 4113-4121, 2016 Characteristics of B cell-associated gene expression in patients with coronary artery disease WENWEN YAN*, HAOMING SONG*, JINFA JIANG, WENJUN XU, ZHU GONG, QIANGLIN DUAN, CHUANGRONG LI, YUAN XIE and LEMIN WANG Department of Internal Medicine, Division of Cardiology, Tongji Hospital, Tongji University School of Medicine, Shanghai 200065, P.R. China Received May 19, 2015; Accepted February 12, 2016 DOI: 10.3892/mmr.2016.5029 Abstract. The current study aimed to identify differentially with the two other groups. Additionally the gene expression expressed B cell-associated genes in peripheral blood mono- levels of B cell regulatory genes were measured. In patients nuclear cells and observe the changes in B cell activation at with AMI, CR1, LILRB2, LILRB3 and VAV1 mRNA expres- different stages of coronary artery disease. Groups of patients sion levels were statistically increased, whereas, CS1 and IL4I1 with acute myocardial infarction (AMI) and stable angina (SA), mRNAs were significantly reduced compared with the SA and as well as healthy volunteers, were recruited into the study control groups. There was no statistically significant difference (n=20 per group). Whole human genome microarray analysis in B cell-associated gene expression levels between patients was performed to examine the expression of B cell-associated with SA and the control group. The present study identified the genes among these three groups. The mRNA expression levels downregulation of genes associated with BCRs, B2 cells and of 60 genes associated with B cell activity and regulation were B cell regulators in patients with AMI, indicating a weakened measured using reverse transcription-quantitative polymerase T cell-B cell interaction and reduced B2 cell activation during chain reaction. -
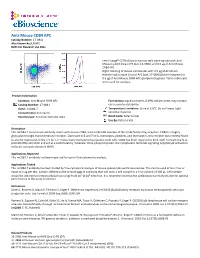
Anti-Mouse CD84 APC Catalog Number: 17‐0841 Also Known As:SLAMF5 RUO: for Research Use Only
Anti-Mouse CD84 APC Catalog Number: 17‐0841 Also Known As:SLAMF5 RUO: For Research Use Only Left: Lineagelo C57BL/6 bone marrow cells were stained with Anti‐ Mouse Ly‐6A/E (Sca‐1) PE (cat. 12‐5981) and 0.5 µg of Anti‐Mouse CD84 APC. Right: Staining of mouse splenocytes with 0.5 µg of Armenian Hamster IgG Isotype Control APC (cat. 17‐4888) (blue histogram) or 0.5 µg of Anti‐Mouse CD84 APC (purple histogram). Total viable cells were used for analysis. Product Information Contents: Anti‐Mouse CD84 APC Formulation: aqueous buffer, 0.09% sodium azide, may contain Catalog Number: 17‐0841 carrier protein/stabilizer Clone: mCD84.7 Temperature Limitation: Store at 2‐8°C. Do not freeze. Light Concentration: 0.2 mg/mL sensitive material. Host/Isotype: Armenian Hamster IgG1 Batch Code: Refer to Vial Use By: Refer to Vial Description This mCD84.7 monoclonal antibody reacts with mouse CD84, a 64‐ to 82‐kDa member of the SLAM family of Ig receptors. CD84 is a highly glycosylated single transmembrane receptor. Expressed in B and T cells, monocytes, platelets, and thymocytes, this receptor was recently found to also be expressed on Sca‐1+c‐kit+Lin‐ mouse bone marrow hematopoietic stem cells. CD84 has been reported to bind itself homophilically to promote IFNγ secretion and act as a costimulatory molecule. Once phosphorylated, the cytoplasmic tail binds signaling lymphocyte activation molecule‐associated protein (SAP). Applications Reported This mCD84.7 antibody has been reported for use in flow cytometric analysis. Applications Tested This mCD84.7 antibody has been tested by flow cytometric analysis of mouse splenocytes and bone marrow.