Potential Silencing of Gene Expression by PIWI-Interacting Rnas (Pirnas) in Somatic Tissues in Mollusk
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Not Dicer but Argonaute Is Required for a Microrna Production
Cell Research (2010) 20:735-737. npg © 2010 IBCB, SIBS, CAS All rights reserved 1001-0602/10 $ 32.00 RESEARCH HIGHLIGHT www.nature.com/cr A new twist in the microRNA pathway: Not Dicer but Argonaute is required for a microRNA production Gabriel D Bossé1, Martin J Simard1 1Laval University Cancer Research Centre, Hôtel-Dieu de Québec (CHUQ), Quebec City, Québec G1R 2J6, Canada Cell Research (2010) 20:735-737. doi:10.1038/cr.2010.83; published online 15 June 2010 Found in all metazoans, microRNAs A Canonical pathway B Ago2-dependent pathway or miRNAs are small non-coding RNA Nucleus Cytoplasm Nucleus Cytoplasm of ~22 nucleotides in length that com- Exp.5 Exp.5 pletely reshaped our understanding of gene regulation. This new class of gene pre-miR-451 regulator is mostly transcribed by the pre-miRNA RNA polymerase II producing a long stem-loop structure, called primary- or Ago2 pri-miRNA, that will first be processed Ago2 Dicer in the cell nucleus by a multiprotein TRBP complex called microprocessor to gen- erate a shorter RNA structure called Ago2 RISC precursor- or pre-miRNA. The precisely Ago2 RISC processed pre-miRNA will next be ex- ported into the cytoplasm by Exportin 5 and loaded onto another processing machine containing the ribonuclease III enzyme Dicer, an Argonaute protein Ago2 Ago2 and other accessory cellular factors mRNA mRNA (Figure 1A; [1]). Dicer will mediate the Translation inhibition Translation inhibition cleavage of the pre-miRNA to form the mature miRNA that will then be bound Figure 1 (A) Canonical microRNA biogenesis. In mammals, the pre-miRNA is by the Argonaute protein to form, most loaded onto a multiprotein complex consisting minimally of Dicer, Tar RNA Bind- likely with other cellular factors, the ef- ing Protein (TRBP) and Ago2. -

Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-Rays-Treated Lymphoblastoid Cells
Int. J. Mol. Sci. 2013, 14, 9099-9110; doi:10.3390/ijms14059099 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-rays-Treated Lymphoblastoid Cells M. Ahmad Chaudhry Department of Medical Laboratory and Radiation Sciences, University of Vermont, Burlington, VT 05405, USA; E-Mail: [email protected]; Tel.: +1-802-656-0569; Fax: +1-802-656-2191 Received: 5 March 2013; in revised version: 19 April 2013 / Accepted: 22 April 2013 / Published: 25 April 2013 Abstract: A wide variety of biological effects are induced in cells that are exposed to ionizing radiation. The expression changes of coding mRNA and non-coding micro-RNA have been implicated in irradiated cells. The involvement of other classes of non-coding RNAs (ncRNA), such as small nucleolar RNAs (snoRNAs), long ncRNAs (lncRNAs), and PIWI-interacting RNAs (piRNAs) in cells recovering from radiation-induced damage has not been examined. Thus, we investigated whether these ncRNA were undergoing changes in cells exposed to ionizing radiation. The modulation of ncRNAs expression was determined in human TK6 (p53 positive) and WTK1 (p53 negative) cells. The snoRNA host genes SNHG1, SNHG6, and SNHG11 were induced in TK6 cells. In WTK1 cells, SNHG1 was induced but SNHG6, and SNHG11 were repressed. SNHG7 was repressed in TK6 cells and was upregulated in WTK1 cells. The lncRNA MALAT1 and SOX2OT were induced in both TK6 and WTK1 cells and SRA1 was induced in TK6 cells only. Interestingly, the MIAT and PIWIL1 were not expressed in TK6 cells before or after the ionizing radiation treatment. -
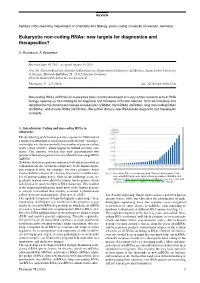
Eukaryotic Non-Coding Rnas: New Targets for Diagnostics and Therapeutics?
REVIEW Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Germany Eukaryotic non-coding RNAs: new targets for diagnostics and therapeutics? O. Rossbach, A. Bindereif Received June 30, 2015, accepted August 19, 2015 Prof. Dr. Albrecht Bindereif, Institute of Biochemistry, Department of Chemistry and Biology, Justus Liebig University of Giessen, Heinrich-Buff-Ring 58, 35392 Giessen, Germany [email protected] Pharmazie 71: 3–7 (2016) doi: 10.1691/ph.2016.5736 Non-coding RNAs (ncRNAs) in eukaryotes have recently developed to a very active research area in RNA biology, opening up new strategies for diagnosis and therapies of human disease. Here we introduce and describe the most important classes of eukaryotic ncRNAs: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). We further discuss new RNA-based diagnostic and therapeutic concepts. 1. Introduction: Coding and non-coding RNAs in eukaryotes The deciphering of the human genome sequence in 2000 marked a unique breakthrough in modern molecular biology. An impor- tant insight was the unexpectedly low number of protein-coding genes (about 20,000), which lagged far behind previous esti- mates. This number revealed that only approximately two percent of the human genome is transcribed into messenger RNA (mRNA). However, the human genome sequence itself did not provide an explanation for the enormous complexity of the human organ- ism compared with, for example, the more primitive worm Caenorhabditis elegans (C. elegans) that carries a similar num- Fig. 1: Non-coding RNAs as an emerging field. The rapid development of the ber of protein-coding genes. -

Epigenetic Roles of PIWI‑Interacting Rnas (Pirnas) in Cancer Metastasis (Review)
ONCOLOGY REPORTS 40: 2423-2434, 2018 Epigenetic roles of PIWI‑interacting RNAs (piRNAs) in cancer metastasis (Review) JIA LIU1, SHUJUN ZHANG2 and BINGLIN CHENG1 1Department of Integrated Traditional Chinese and Western Medicine Oncology, The First Affiliated Hospital of Harbin Medical University; 2Department of Pathology, The Fourth Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150000, P.R. China Received March 19, 2018; Accepted September 3, 2018 DOI: 10.3892/or.2018.6684 Abstract. P-element-induced wimpy testis (PIWI)-interacting 7. Epigenetics of ncRNAs in cancer RNAs (piRNAs) are epigenetic-related short ncRNAs that 8. Discussion participate in chromatin regulation, transposon silencing, and modification of specific gene sites. These epigenetic factors or alterations are also involved in the growth of a variety 1. Introduction of human cancers, including lung, breast, and colon cancer. Accumulating evidence has revealed that tumor metastasis P-element-induced wimpy testis (PIWI)-interacting RNAs and invasion involve genetic and epigenetic factors. Cancer (piRNAs) belong to a new class of ncRNAs that have been asso- metastasis is characterized by epigenetic alterations including ciated with many cancers (1). piRNAs are involved in the gene DNA methylation and histone modification. Changes in DNA regulation process in which certain nucleotides bind coding methylation, H3K9me3 heterochromatin and transposable regions in gene promoters (2). piRNAs function in the epigen- elements have been detected in several cancers. piRNAs may etic regulation of DNA methylation (3), transposable silencing function in gene silencing and gene modification upstream and chromatin modification (4). PIWI is a type of Argonaute or downstream of oncogenes in cancer cell lines or cancer protein that binds to piRNAs and carries out unique functions tissues. -

RNA Interference in the Nucleus: Roles for Small Rnas in Transcription, Epigenetics and Beyond
REVIEWS NON-CODING RNA RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond Stephane E. Castel1 and Robert A. Martienssen1,2 Abstract | A growing number of functions are emerging for RNA interference (RNAi) in the nucleus, in addition to well-characterized roles in post-transcriptional gene silencing in the cytoplasm. Epigenetic modifications directed by small RNAs have been shown to cause transcriptional repression in plants, fungi and animals. Additionally, increasing evidence indicates that RNAi regulates transcription through interaction with transcriptional machinery. Nuclear small RNAs include small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs) and are implicated in nuclear processes such as transposon regulation, heterochromatin formation, developmental gene regulation and genome stability. RNA interference Since the discovery that double-stranded RNAs (dsRNAs) have revealed a conserved nuclear role for RNAi in (RNAi). Silencing at both the can robustly silence genes in Caenorhabditis elegans and transcriptional gene silencing (TGS). Because it occurs post-transcriptional and plants, RNA interference (RNAi) has become a new para- in the germ line, TGS can lead to transgenerational transcriptional levels that is digm for understanding gene regulation. The mecha- inheritance in the absence of the initiating RNA, but directed by small RNA molecules. nism is well-conserved across model organisms and it is dependent on endogenously produced small RNA. uses short antisense RNA to inhibit translation or to Such epigenetic inheritance is familiar in plants but has Post-transcriptional gene degrade cytoplasmic mRNA by post-transcriptional gene only recently been described in metazoans. silencing silencing (PTGS). PTGS protects against viral infection, In this Review, we cover the broad range of nuclear (PTGS). -

RNA Stem Structure Governs Coupling of Dicing and Gene Silencing in RNA
RNA stem structure governs coupling of dicing and PNAS PLUS gene silencing in RNA interference Hye Ran Koha,b,1, Amirhossein Ghanbariniakia,c,d, and Sua Myonga,c,d,1 aDepartment of Biophysics, Johns Hopkins University, Baltimore, MD 21218; bDepartment of Chemistry, Chung-Ang University, Seoul 06974, Korea; cInstitute for Genomic Biology, University of Illinois, Urbana, IL 61801; and dCenter for Physics of Living Cells, University of Illinois, Urbana, IL 61801 Edited by Brenda L. Bass, University of Utah School of Medicine, Salt Lake City, UT, and approved October 13, 2017 (received for review June 8, 2017) PremicroRNAs (premiRNAs) possess secondary structures consist- coworkers (20), but its relationship to the silencing efficiency has ing of a loop and a stem with multiple mismatches. Despite the not been tested. While the effect of mismatches between the well-characterized RNAi pathway, how the structural features of guide strand and target mRNA has been extensively studied (21– premiRNA contribute to dicing and subsequent gene-silencing 24), the effect of mismatch between guide and passenger strand efficiency remains unclear. Using single-molecule FISH, we dem- has not been systematically examined. onstrate that cytoplasmic mRNA, but not nuclear mRNA, is reduced We sought to study how different structural features found in during RNAi. The dicing rate and silencing efficiency both increase premiRNAs and presiRNAs modulate overall gene-silencing in a correlated manner as a function of the loop length. In contrast, efficiency by measuring two key steps in the RNAi pathway: (i) mismatches in the stem drastically diminish the silencing efficiency dicing kinetics to measure how quickly premiRNA or presiRNA without impacting the dicing rate. -

A Pirna-Like Small RNA Interacts with and Modulates P-ERM Proteins in Human Somatic Cells
ARTICLE Received 18 Mar 2015 | Accepted 27 Apr 2015 | Published 22 June 2015 DOI: 10.1038/ncomms8316 OPEN A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells Yuping Mei1, Yuyan Wang1,2, Priti Kumari3, Amol Carl Shetty3, David Clark1, Tyler Gable1, Alexander D. MacKerell4,5, Mark Z. Ma1, David J. Weber5,6, Austin J. Yang5, Martin J. Edelman5 & Li Mao1,5 PIWI-interacting RNAs (piRNAs) are thought to silence transposon and gene expression during development. However, the roles of piRNAs in somatic tissues are largely unknown. Here we report the identification of 555 piRNAs in human lung bronchial epithelial (HBE) and non-small cell lung cancer (NSCLC) cell lines, including 295 that do not exist in databases termed as piRNA-like sncRNAs or piRNA-Ls. Distinctive piRNA/piRNA-L expression patterns are observed between HBE and NSCLC cells. piRNA-like-163 (piR-L-163), the top down- regulated piRNA-L in NSCLC cells, binds directly to phosphorylated ERM proteins (p-ERM), which is dependent on the central part of UUNNUUUNNUU motif in piR-L-163 and the RRRKPDT element in ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein 50 (EBP50). Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions. 1 Department of Oncology and Diagnostic Sciences, University of Maryland School of Dentistry, 650W Baltimore Street, Baltimore, Maryland 21201, USA. 2 Department of Thoracic Medical Oncology, Peking University Cancer Hospital and Institute, Beijing 100142, China. -

Xenopus Piwi Proteins Interact with a Broad Proportion of the Oocyte Transcriptome
Downloaded from rnajournal.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome JAMES A. TOOMBS,1,2,4 YULIYA A. SYTNIKOVA,3,5 GUNG-WEI CHIRN,3,6 IGNATIUS ANG,3,7 NELSON C. LAU,3 and MICHAEL D. BLOWER1,2 1Department of Molecular Biology, Massachusetts General Hospital, Boston, Massachusetts 02114, USA 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA 3Department of Biology and Rosenstiel Basic Medical Science Research Center, Brandeis University, Waltham, Massachusetts 02454, USA ABSTRACT Piwi proteins utilize small RNAs (piRNAs) to recognize target transcripts such as transposable elements (TE). However, extensive piRNA sequence diversity also suggests that Piwi/piRNA complexes interact with many transcripts beyond TEs. To determine Piwi target RNAs, we used ribonucleoprotein-immunoprecipitation (RIP) and cross-linking and immunoprecipitation (CLIP) to identify thousands of transcripts associated with the Piwi proteins XIWI and XILI (Piwi-protein-associated transcripts, PATs) from early stage oocytes of X. laevis and X. tropicalis. Most PATs associate with both XIWI and XILI and include transcripts of developmentally important proteins in oogenesis and embryogenesis. Only a minor fraction of PATs in both frog species displayed near perfect matches to piRNAs. Since predicting imperfect pairing between all piRNAs and target RNAs remains intractable, we instead determined that PAT read counts correlate well with the lengths and expression levels of transcripts, features that have also been observed for oocyte mRNAs associated with Drosophila Piwi proteins. We used an in vitro assay with exogenous RNA to confirm that XIWI associates with RNAs in a length- and concentration-dependent manner. -

INTRODUCTION Sirna and Rnai
J Korean Med Sci 2003; 18: 309-18 Copyright The Korean Academy ISSN 1011-8934 of Medical Sciences RNA interference (RNAi) is the sequence-specific gene silencing induced by dou- ble-stranded RNA (dsRNA). Being a highly specific and efficient knockdown tech- nique, RNAi not only provides a powerful tool for functional genomics but also holds Institute of Molecular Biology and Genetics and School of Biological Science, Seoul National a promise for gene therapy. The key player in RNAi is small RNA (~22-nt) termed University, Seoul, Korea siRNA. Small RNAs are involved not only in RNAi but also in basic cellular pro- cesses, such as developmental control and heterochromatin formation. The inter- Received : 19 May 2003 esting biology as well as the remarkable technical value has been drawing wide- Accepted : 23 May 2003 spread attention to this exciting new field. V. Narry Kim, D.Phil. Institute of Molecular Biology and Genetics and School of Biological Science, Seoul National University, San 56-1, Shillim-dong, Gwanak-gu, Seoul 151-742, Korea Key Words : RNA Interference (RNAi); RNA, Small interfering (siRNA); MicroRNAs (miRNA); Small Tel : +82.2-887-8734, Fax : +82.2-875-0907 hairpin RNA (shRNA); mRNA degradation; Translation; Functional genomics; Gene therapy E-mail : [email protected] INTRODUCTION established yet, testing 3-4 candidates are usually sufficient to find effective molecules. Technical expertise accumulated The RNA interference (RNAi) pathway was originally re- in the field of antisense oligonucleotide and ribozyme is now cognized in Caenorhabditis elegans as a response to double- being quickly applied to RNAi, rapidly improving RNAi stranded RNA (dsRNA) leading to sequence-specific gene techniques. -

Piwi-Interacting Rnas and PIWI Genes As Novel Prognostic Markers for Breast Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 7, No. 25 Research Paper Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer Preethi Krishnan1, Sunita Ghosh2,3, Kathryn Graham2,3, John R. Mackey2,3, Olga Kovalchuk4, Sambasivarao Damaraju1,3 1Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta, Canada 2Department of Oncology, University of Alberta, Edmonton, Alberta, Canada 3Cross Cancer Institute, Alberta Health Services, Edmonton, Alberta, Canada 4Department of Biological Sciences, University of Lethbridge, Lethbridge, Alberta, Canada Correspondence to: Sambasivarao Damaraju, email: [email protected] Keywords: piRNA, PIWI, breast cancer, prognostic marker, TCGA Received: January 13, 2016 Accepted: April 28, 2016 Published: May 10, 2016 ABSTRACT Piwi-interacting RNAs (piRNAs), whose role in germline maintenance has been established, are now also being classified as post-transcriptional regulators of gene expression in somatic cells. PIWI proteins, central to piRNA biogenesis, have been identified as genetic and epigenetic regulators of gene expression. piRNAs/PIWIs have emerged as potential biomarkers for cancer but their relevance to breast cancer has not been comprehensively studied. piRNAs and mRNAs were profiled from normal and breast tumor tissues using next generation sequencing and Agilent platforms, respectively. Gene targets for differentially expressed piRNAs were identified from mRNA expression dataset. piRNAs and PIWI genes were independently assessed for their prognostic significance (outcomes: Overall Survival, OS and Recurrence Free Survival, RFS). We discovered eight piRNAs as novel independent prognostic markers and their association with OS was confirmed in an external dataset (The Cancer Genome Atlas). Further, PIWIL3 and PIWIL4 genes showed prognostic relevance. 306 gene targets exhibited reciprocal relationship with piRNA expression. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

Questions with Answers- Nucleotides & Nucleic Acids A. the Components
Questions with Answers- Nucleotides & Nucleic Acids A. The components and structures of common nucleotides are compared. (Questions 1-5) 1._____ Which structural feature is shared by both uracil and thymine? a) Both contain two keto groups. b) Both contain one methyl group. c) Both contain a five-membered ring. d) Both contain three nitrogen atoms. 2._____ Which component is found in both adenosine and deoxycytidine? a) Both contain a pyranose. b) Both contain a 1,1’-N-glycosidic bond. c) Both contain a pyrimidine. d) Both contain a 3’-OH group. 3._____ Which property is shared by both GDP and AMP? a) Both contain the same charge at neutral pH. b) Both contain the same number of phosphate groups. c) Both contain the same purine. d) Both contain the same furanose. 4._____ Which characteristic is shared by purines and pyrimidines? a) Both contain two heterocyclic rings with aromatic character. b) Both can form multiple non-covalent hydrogen bonds. c) Both exist in planar configurations with a hemiacetal linkage. d) Both exist as neutral zwitterions under cellular conditions. 5._____ Which property is found in nucleosides and nucleotides? a) Both contain a nitrogenous base, a pentose, and at least one phosphate group. b) Both contain a covalent phosphodister bond that is broken in strong acid. c) Both contain an anomeric carbon atom that is part of a β-N-glycosidic bond. d) Both contain an aldose with hydroxyl groups that can tautomerize. ___________________________________________________________________________ B. The structures of nucleotides and their components are studied. (Questions 6-10) 6._____ Which characteristic is shared by both adenine and cytosine? a) Both contain one methyl group.