Multiple Multidimensional Sequence Alignment Using Generalized
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Logmine: Fast Pattern Recognition for Log Analytics
LogMine: Fast Pattern Recognition for Log Analytics Hossein Hamooni Biplob Debnath Jianwu Xu University of New Mexico NEC Laboratories America NEC Laboratories America 1 University Blvd 4 Independence Way 4 Independence Way Albuquerque, NM 87131 Princeton, NJ 08540 Princeton, NJ 08540 [email protected] [email protected] [email protected] Hui Zhang Guofei Jiang Abdullah Mueen NEC Laboratories America NEC Laboratories America University of New Mexico 4 Independence Way 4 Independence Way 1 University Blvd Princeton, NJ 08540 Princeton, NJ 08540 Albuquerque, NM 87131 [email protected] [email protected] [email protected] ABSTRACT Keywords Modern engineering incorporates smart technologies in all Log analysis; Pattern recognition; Map-reduce aspects of our lives. Smart technologies are generating ter- abytes of log messages every day to report their status. It 1. INTRODUCTION is crucial to analyze these log messages and present usable The Internet of Things (IoT) enables advanced connec- information (e.g. patterns) to administrators, so that they tivity of computing and embedded devices through internet can manage and monitor these technologies. Patterns mini- infrastructure. Although computers and smartphones are mally represent large groups of log messages and enable the the most common devices in IoT, the number of \things" administrators to do further analysis, such as anomaly de- is expected to grow to 50 billion by 2020 [5]. IoT involves tection and event prediction. Although patterns exist com- machine-to-machine communications (M2M), where it is im- monly in automated log messages, recognizing them in mas- portant to continuously monitor connected machines to de- sive set of log messages from heterogeneous sources without tect any anomaly or bug, and resolve them quickly to mini- any prior information is a significant undertaking. -

Role of Similarity Measures in Time Series Analysis
UNIVERSITY OF NOVI SAD FACULTY OF SCIENCE DEPARTMENT OF MATHEMATICS AND INFORMATICS Zoltan Geler Role of Similarity Measures in Time Series Analysis - Doctoral Dissertation - Mentor: Prof. Dr. Mirjana Ivanović Novi Sad, 2015 Preface The main subject of this dissertation is a comprehensive examination of the role of similarity measures in time-series analysis through studying the influence of the Sakoe-Chiba band on accuracy of classification. A time series represents a series of numerical data points in successive order, usually with uniform intervals between them. Time series are used for storage, analysis and visualization of data collected in many different domains, including science, medicine, economics and others. The increasing demand to work with large amounts of data has led to a growing interest in researching different tasks of time-series data mining. In recent years, there is an increasing interest in studying various aspects of time-series classification. One of the most important issues of time-series classification is a thoughtful and adequate choice of the similarity measure. They enable comparison of time series by describing their similarity (or dissimilarity) with numerical values. In the field of time-series data mining a great number of different similarity measures has been proposed and used. Many of these measures are implemented using dynamic programming. Unfortunately, the time requirements of this technique may adversely affect the possibilities of its application in practice. One way of dealing with this issue is to limit the search area by applying global constraints. However, these restrictions may affect the accuracy of classification, and therefore understanding their impact is of great importance. -

Similarity Processing in Multi-Observation Data
Similarity Processing in Multi-Observation Data Thomas Bernecker Munchen¨ 2012 Similarity Processing in Multi-Observation Data Thomas Bernecker Dissertation an der Fakult¨at fur¨ Mathematik, Informatik und Statistik der Ludwig{Maximilians{Universit¨at Munchen¨ vorgelegt von Thomas Bernecker aus Munchen¨ Munchen,¨ den 24.08.2012 Erstgutachter: Prof. Dr. Hans-Peter Kriegel, Ludwig-Maximilians-Universit¨at Munchen¨ Zweitgutachter: Prof. Mario Nascimento, University of Alberta Tag der mundlichen¨ Prufung:¨ 21.12.2012 v Abstract Many real-world application domains such as sensor-monitoring systems for environmental research or medical diagnostic systems are dealing with data that is represented by multiple observations. In contrast to single-observation data, where each object is assigned to exactly one occurrence, multi-observation data is based on several occurrences that are subject to two key properties: temporal variability and uncertainty. When defining similarity between data objects, these properties play a significant role. In general, methods designed for single- observation data hardly apply for multi-observation data, as they are either not supported by the data models or do not provide sufficiently efficient or effective solutions. Prominent directions incorporating the key properties are the fields of time series, where data is created by temporally successive observations, and uncertain data, where observations are mutually exclusive. This thesis provides research contributions for similarity processing { similarity search and data mining { on time series and uncertain data. The first part of this thesis focuses on similarity processing in time series databases. A variety of similarity measures have recently been proposed that support similarity pro- cessing w.r.t. various aspects. In particular, this part deals with time series that consist of periodic occurrences of patterns. -

Clustering Human Behaviors with Dynamic Time Warping and Hidden Markov Models for a Video Surveillance System
Clustering Human Behaviors with Dynamic Time Warping and Hidden Markov Models for a Video Surveillance System Kan Ouivirach and Matthew N. Dailey Computer Science and Information Management Asian Institute of Technology [email protected], [email protected] Abstract—We propose and experimentally evaluate a new by constructing an affinity matrix using dynamic time warping method for clustering human behaviors that is suitable for (DTW) [7] then apply the normalized-cut approach to cluster bootstrapping an anomaly detection module for intelligent video the gestures. Hautamaki¨ et al. [8] apply DTW and use the surveillance systems. The method uses dynamic time warping, ag- glomerative hierarchical clustering, and hidden Markov models pairwise DTW distances as input to a hierarchical clustering to provide an initial partitioning of a set of observation sequences process in which k-means is used to fine-tune the output. then automatically identifies where to cut off the hierarchical In this paper, we use a combination of clustering and HMMs clustering dendrogram. We show that the method is extremely to group human behaviors in a scene. There is some recent effective, providing 100% accuracy in separating anomalous from related work using graphical models such as HMMs to cluster typical behaviors on real-world testbed video surveillance data. behavior patterns. Li and Biswas [9] use the Bayesian informa- tion criterion (BIC) for HMM model selection and construct I. INTRODUCTION a binary hierarchical clustering dendrogram to initialize data Human behavior understanding is an important component partitions based on a sequence-to-model likelihood distance of a wide variety of desirable intelligent systems. -

Diffeomorphic Temporal Alignment Nets
Diffeomorphic Temporal Alignment Nets Ron Shapira Weber Matan Eyal Ben-Gurion University Ben-Gurion University [email protected] [email protected] Nicki Skafte Detlefsen Oren Shriki Oren Freifeld Technical University of Denmark Ben-Gurion University Ben-Gurion University [email protected] [email protected] [email protected] Abstract Time-series analysis is confounded by nonlinear time warping of the data. Tradi- tional methods for joint alignment do not generalize: after aligning a given signal ensemble, they lack a mechanism, that does not require solving a new optimization problem, to align previously-unseen signals. In the multi-class case, they must also first classify the test data before aligning it. Here we propose the Diffeomorphic Temporal Alignment Net (DTAN), a learning-based method for time-series joint alignment. Via flexible temporal transformer layers, DTAN learns and applies an input-dependent nonlinear time warping to its input signal. Once learned, DTAN easily aligns previously-unseen signals by its inexpensive forward pass. In a single- class case, the method is unsupervised: the ground-truth alignments are unknown. In the multi-class case, it is semi-supervised in the sense that class labels (but not the ground-truth alignments) are used during learning; in test time, however, the class labels are unknown. As we show, DTAN not only outperforms existing joint- alignment methods in aligning training data but also generalizes well to test data. Our code is available at https://github.com/BGU-CS-VIL/dtan. 1 Introduction Time-series data often presents a significant amount of misalignment, also known as nonlinear time warping. -

Eventdtw: an Improved Dynamic Time Warping Algorithm for Aligning Biomedical Signals of Nonuniform Sampling Frequencies
sensors Article EventDTW: An Improved Dynamic Time Warping Algorithm for Aligning Biomedical Signals of Nonuniform Sampling Frequencies 1, 1, 1 1 2 Yihang Jiang y, Yuankai Qi y, Will Ke Wang , Brinnae Bent , Robert Avram , Jeffrey Olgin 2 and Jessilyn Dunn 1,* 1 The Departments of Biomedical Engineering and Biostatistics & Bioinformatics, Duke University, Durham, NC 27708, USA; [email protected] (Y.J.); [email protected] (Y.Q.); [email protected] (W.K.W.); [email protected] (B.B.) 2 The Division of Cardiology and the Cardiovascular Research Institute, University of California San Francisco, San Francisco, CA 94143, USA; [email protected] (R.A.); jeff[email protected] (J.O.) * Correspondence: [email protected] First co-authors. y Received: 19 March 2020; Accepted: 6 May 2020; Published: 9 May 2020 Abstract: The dynamic time warping (DTW) algorithm is widely used in pattern matching and sequence alignment tasks, including speech recognition and time series clustering. However, DTW algorithms perform poorly when aligning sequences of uneven sampling frequencies. This makes it difficult to apply DTW to practical problems, such as aligning signals that are recorded simultaneously by sensors with different, uneven, and dynamic sampling frequencies. As multi-modal sensing technologies become increasingly popular, it is necessary to develop methods for high quality alignment of such signals. Here we propose a DTW algorithm called EventDTW which uses information propagated from defined events as basis for path matching and hence sequence alignment. We have developed two metrics, the error rate (ER) and the singularity score (SS), to define and evaluate alignment quality and to enable comparison of performance across DTW algorithms. -

Shape Dynamic Time Warping
Pattern Recognition 74 (2018) 171–184 Contents lists available at ScienceDirect Pattern Recognition journal homepage: www.elsevier.com/locate/patcog shapeDTW: Shape Dynamic Time Warping ∗ Jiaping Zhao , Laurent Itti University of Southern California, USA a r t i c l e i n f o a b s t r a c t Article history: Dynamic Time Warping (DTW) is an algorithm to align temporal sequences with possible local non-linear Received 17 October 2016 distortions, and has been widely applied to audio, video and graphics data alignments. DTW is essentially Revised 5 August 2017 a point-to-point matching method under some boundary and temporal consistency constraints. Although Accepted 12 September 2017 DTW obtains a global optimal solution, it does not necessarily achieve locally sensible matchings. Con- Available online 14 September 2017 cretely, two temporal points with entirely dissimilar local structures may be matched by DTW. To ad- Keywords: dress this problem, we propose an improved alignment algorithm, named shape Dynamic Time Warping Dynamic Time Warping (shapeDTW), which enhances DTW by taking point-wise local structural information into consideration. Sequence alignment shapeDTW is inherently a DTW algorithm, but additionally attempts to pair locally similar structures Time series classification and to avoid matching points with distinct neighborhood structures. We apply shapeDTW to align audio signal pairs having ground-truth alignments, as well as artificially simulated pairs of aligned sequences, and obtain quantitatively much lower alignment errors than DTW and its two variants. When shapeDTW is used as a distance measure in a nearest neighbor classifier (NN-shapeDTW) to classify time series, it beats DTW on 64 out of 84 UCR time series datasets, with significantly improved classification accuracies. -
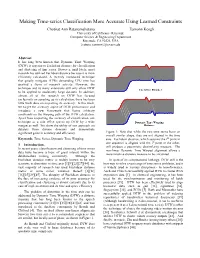
Making Time-Series Classification More Accurate Using Learned
Making Time-series Classification More Accurate Using Learned Constraints Chotirat Ann Ratanamahatana Eamonn Keogh University of California - Riverside Computer Science & Engineering Department Riverside, CA 92521, USA {ratana, eamonn}@cs.ucr.edu Abstract It has long been known that Dynamic Time Warping (DTW) is superior to Euclidean distance for classification and clustering of time series. However, until lately, most research has utilized Euclidean distance because it is more efficiently calculated. A recently introduced technique that greatly mitigates DTWs demanding CPU time has sparked a flurry of research activity. However, the technique and its many extensions still only allow DTW Euclidean Distance to be applied to moderately large datasets. In addition, almost all of the research on DTW has focused exclusively on speeding up its calculation; there has been little work done on improving its accuracy. In this work, we target the accuracy aspect of DTW performance and introduce a new framework that learns arbitrary constraints on the warping path of the DTW calculation. Apart from improving the accuracy of classification, our technique as a side effect speeds up DTW by a wide Dynamic Time Warping margin as well. We show the utility of our approach on Distance datasets from diverse domains and demonstrate significant gains in accuracy and efficiency. Figure 1: Note that while the two time series have an overall similar shape, they are not aligned in the time Keywords: Time Series, Dynamic Time Warping. axis. Euclidean distance, which assumes the ith point in one sequence is aligned with the ith point in the other, 1 Introduction. will produce a pessimistic dissimilarity measure. -

Mining and Similarity Search in Temporal Databases
1 ERGEBNISSE AUS DER INFORMATIK Insights from database research, notably in the areas of data mining and similarity search, and advances in storage and microprocessor technology have enabled users to analyze and explore large-scale datasets. Data mining is the task of extracting previously unknown knowledge from data; similarity search encompasses techniques for finding objects similar by content. A prominent kind of data used in these tasks are temporal datasets, which stand out due to their information richness and their many possible applications. This thesis contributes novel, advanced methods for data mining and simi- larity search on temporal databases. Hardy Kremer A major challenge in data mining research is the effectiveness of the approaches, corre- sponding to the quality of extracted patterns. The thesis addresses this challenge for the mining task of temporal clustering. First, a clustering technique is developed that is spe- cifically designed for the requirements of real world time series. Even in difficult settings with various measurement errors and misalignments between time series, it correctly identifies patterns concealed in temporal or dimensional subspaces of the data domain. Mining and Similarity Search Second, new methods for the complex task of mapping clusters between clusterings are contributed, for which two applications are investigated: tracing of evolving clusters in in Temporal Databases spatio-temporal data and the evaluation of clustering results in data stream scenarios. The core of content-based similarity search systems and many data mining tasks are distance functions measuring the similarity between objects. An effective but also com- putationally expensive distance function for time series is based on adaptive warping on the time axis. -

Comparison of HMM and DTW for Isolated Word Recognition System of Punjabi Language
Comparison of HMM and DTW for Isolated Word Recognition System of Punjabi Language Kumar Ravinder Department of Computer Science & Engineering, Thapar Univeristy, Patiala – 147004 (India) [email protected] Abstract. Issue of speech interface to computer has been capturing the global attention because of convenience put forth by it. Although speech recognition is not a new phenomenon in existing developments of user-machine interface studies but the highlighted facts only provide promising solutions for widely accepted language English. This paper presents development of an experimen- tal, speaker-dependent, real-time, isolated word recognizer for Indian regional language Punjabi. Research is further extended to comparison of speech recog- nition system for small vocabulary of speaker dependent isolated spoken words in Indian regional language (Punjabi) using the Hidden Markov Model (HMM) and Dynamic Time Warp (DTW) technique. Punjabi language gives immense changes between consecutive phonemes. Thus, end point detection becomes highly difficult. The presented work emphasizes on template-based recognizer approach using linear predictive coding with dynamic programming computa- tion and vector quantization with Hidden Markov Model based recognizers in isolated word recognition tasks, which also significantly reduces the computa- tional costs. The parametric variation gives enhancement in the feature vector for recognition of 500-isolated word vocabulary on Punjabi language, as the Hidden Marko Model and Dynamic Time Warp technique gives 91.3% and 94.0% accuracy respectively. Keywords: Dynamic programming (DP), dynamic time warp (DTW), hidden markov model (HMM), linear predictive coding (LPC), Punjabi language, vec- tor quantization (VQ). 1 Introduction Research in automatic speech recognition has been known for many years, various researchers have tried to analyze the different aspects of speech in Indian languages. -
Needleman–Wunsch Algorithm
Not logged in Talk Contributions Create account Log in Article Talk Read Edit View history Search Wikipedia Wiki Loves Love: Documenting festivals and celebrations of love on Commons. Help Wikimedia and win prizes by sending photos. Main page Contents Featured content Current events Needleman–Wunsch algorithm Random article From Wikipedia, the free encyclopedia Donate to Wikipedia Wikipedia store This article may be too technical for most readers to understand. Please help improve it Interaction to make it understandable to non-experts, without removing the technical details. (September 2013) (Learn how and when to remove this template message) Help About Wikipedia The Needleman–Wunsch algorithm is an algorithm used in bioinformatics to align Class Sequence Community portal protein or nucleotide sequences. It was one of the first applications of dynamic alignment Recent changes Worst-case performance Contact page programming to compare biological sequences. The algorithm was developed by Saul B. Needleman and Christian D. Wunsch and published in 1970.[1] The Worst-case space complexity Tools algorithm essentially divides a large problem (e.g. the full sequence) into a series of What links here smaller problems and uses the solutions to the smaller problems to reconstruct a solution to the larger problem.[2] It is also Related changes sometimes referred to as the optimal matching algorithm and the global alignment technique. The Needleman–Wunsch algorithm Upload file is still widely used for optimal global alignment, particularly when -

Neural Time Warping for Multiple Sequence Alignment
Neural Time Warping For Multiple Sequence Alignment Keisuke Kawano, Takuro Kutsuna, Satoshi Koide Toyota Central R&D Labs. Inc., Nagakute, Aichi, Japan Abstract Multiple sequences alignment (MSA) is a traditional and challenging task for time-series analyses. The MSA problem is formulated as a discrete optimization problem and is typically solved by dynamic programming. However, the computational complexity increases exponentially with respect to the number of input sequences. In this paper, we propose neural time warping (NTW) that relaxes the original MSA to a continuous optimization and obtains the alignments using a neural network. The solution obtained by NTW is guaranteed to be a feasible solution for the original discrete optimization problem under mild conditions. Our experimental results show that NTW successfully aligns a hundred time-series and significantly outperforms existing methods for solving the MSA problem. In addition, we show a method for obtaining average time-series data as one of applications of NTW. Compared to the existing barycenters, the mean time series data retains the features of the input time-series data. 1 Introduction Time-series alignment is one of the most fundamental operations in processing sequential data, and has various applications including speech recognition [1], computer vision [2, 3], and human activity analysis [4, 5]. Alignment can be written as a discrete optimization problem that determines how to warp each sequence to match the timing of patterns in the sequences. Dynamic time warping (DTW) [6] is a prominent approach, which finds the optimal alignment using the dynamic programming technique. However, for multiple sequence alignment (MSA), it is difficult to utilize DTW because the computational complexity increases exponentially with the number of time-series data.