Nearest Neighbor Methods Philip M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

User Guide April 2004 EPA/600/R04/079 April 2004
ProUCL Version 3.0 User Guide April 2004 EPA/600/R04/079 April 2004 ProUCL Version 3.0 User Guide by Anita Singh Lockheed Martin Environmental Services 1050 E. Flamingo Road, Suite E120, Las Vegas, NV 89119 Ashok K. Singh Department of Mathematical Sciences University of Nevada, Las Vegas, NV 89154 Robert W. Maichle Lockheed Martin Environmental Services 1050 E. Flamingo Road, Suite E120, Las Vegas, NV 89119 i Table of Contents Authors ..................................................................... i Table of Contents ............................................................. ii Disclaimer ................................................................. vii Executive Summary ........................................................ viii Introduction ................................................................ ix Installation Instructions ........................................................ 1 Minimum Hardware Requirements ............................................... 1 A. ProUCL Menu Structure .................................................... 2 1. File ............................................................... 3 2. View .............................................................. 4 3. Help .............................................................. 5 B. ProUCL Components ....................................................... 6 1. File ................................................................7 a. Input File Format ...............................................9 b. Result of Opening an Input Data File -

Basic Statistical Concepts Statistical Population
Basic Statistical Concepts Statistical Population • The entire underlying set of observations from which samples are drawn. – Philosophical meaning: all observations that could ever be taken for range of inference • e.g. all barnacle populations that have ever existed, that exist or that will exist – Practical meaning: all observations within a reasonable range of inference • e.g. barnacle populations on that stretch of coast 1 Statistical Sample •A representative subset of a population. – What counts as being representative • Unbiased and hopefully precise Strategies • Define survey objectives: what is the goal of survey or experiment? What are your hypotheses? • Define population parameters to estimate (e.g. number of individuals, growth, color etc). • Implement sampling strategy – measure every individual (think of implications in terms of cost, time, practicality especially if destructive) – measure a representative portion of the population (a sample) 2 Sampling • Goal: – Every unit and combination of units in the population (of interest) has an equal chance of selection. • This is a fundamental assumption in all estimation procedures •How: – Many ways if underlying distribution is not uniform » In the absence of information about underlying distribution the only safe strategy is random sampling » Costs: sometimes difficult, and may lead to its own source of bias (if sample size is low). Much more about this later Sampling Objectives • To obtain an unbiased estimate of a population mean • To assess the precision of the estimate (i.e. -

Experimental Investigation of the Tension and Compression Creep Behavior of Alumina-Spinel Refractories at High Temperatures
ceramics Article Experimental Investigation of the Tension and Compression Creep Behavior of Alumina-Spinel Refractories at High Temperatures Lucas Teixeira 1,* , Soheil Samadi 2 , Jean Gillibert 1 , Shengli Jin 2 , Thomas Sayet 1 , Dietmar Gruber 2 and Eric Blond 1 1 Université d’Orléans, Université de Tours, INSA-CVL, LaMé, 45072 Orléans, France; [email protected] (J.G.); [email protected] (T.S.); [email protected] (E.B.) 2 Chair of Ceramics, Montanuniversität Leoben, 8700 Leoben, Austria; [email protected] (S.S.); [email protected] (S.J.); [email protected] (D.G.) * Correspondence: [email protected] Received: 3 September 2020; Accepted: 18 September 2020; Published: 22 September 2020 Abstract: Refractory materials are subjected to thermomechanical loads during their working life, and consequent creep strain and stress relaxation are often observed. In this work, the asymmetric high temperature primary and secondary creep behavior of a material used in the working lining of steel ladles is characterized, using uniaxial tension and compression creep tests and an inverse identification procedure to calculate the parameters of a Norton-Bailey based law. The experimental creep curves are presented, as well as the curves resulting from the identified parameters, and a statistical analysis is made to evaluate the confidence of the results. Keywords: refractories; creep; parameters identification 1. Introduction Refractory materials, known for their physical and chemical stability, are used in high temperature processes in different industries, such as iron and steel making, cement and aerospace. These materials are exposed to thermomechanical loads, corrosion/erosion from solids, liquids and gases, gas diffusion, and mechanical abrasion [1]. -

Introduction to Basic Concepts
Introduction to Basic Concepts ! There are 5 basic steps to resource management: " 1st - Assess current situation (gather and analyze info) " 2nd - Develop a plan " 3rd- Implement the plan " 4th- Monitor results (effects) of plan " 5th- Replan ! This class will help you do the 1st and 4th steps in the process ! For any study of vegetation there are basically 8 steps: " 1st Set objectives - What do you wan to know? You can’t study everything, you must have a goal " 2nd Choose variable you are going to measure Make sure you are meeting your objectives " 3rd Read published research & ask around Find out what is known about your topic and research area " 4th Choose a sampling method, number of samples, analysis technique, and statistical hypothesis " 5th Go collect data - Details, details, details.... Pay attention to detail - Practice and train with selected methods before you start - Be able to adjust once you get to the field " 6th Analyze the data - Summarize data - Use a statistical comparison " 7th Interpret the results - What did you find? Why is it important? - What can your results tell you about the ecosystem? - Remember the limitations of your technique " 8th Write the report - Your results are worthless if they stay in your files - Clearly document your methods - Use tables and figures to explain your data - Be correct and concise - No matter how smart you are, you will sound stupid if you can’t clearly communicate your results A good reference for several aspects of rangeland inventory and monitoring is: http://rangelandswest.org/az/monitoringtechnical.html -

Shapiro, A., “Statistical Inference of Moment Structures”
Statistical Inference of Moment Structures Alexander Shapiro School of Industrial and Systems Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332-0205, USA e-mail: [email protected] Abstract This chapter is focused on statistical inference of moment structures models. Although the theory is presented in terms of general moment structures, the main emphasis is on the analysis of covariance structures. We discuss identifiability and the minimum discrepancy function (MDF) approach to statistical analysis (estimation) of such models. We address such topics of the large samples theory as consistency, asymptotic normality of MDF estima- tors and asymptotic chi-squaredness of MDF test statistics. Finally, we discuss asymptotic robustness of the normal theory based MDF statistical inference in the analysis of covariance structures. 1 Contents 1 Introduction 1 2 Moment Structures Models 1 3 Discrepancy Functions Estimation Approach 6 4 Consistency of MDF Estimators 9 5 Asymptotic Analysis of the MDF Estimation Procedure 12 5.1 Asymptotics of MDF Test Statistics . 15 5.2 Nested Models . 19 5.3 Asymptotics of MDF Estimators . 20 5.4 The Case of Boundary Population Value . 22 6 Asymptotic Robustness of the MDF Statistical Inference 24 6.1 Elliptical distributions . 26 6.2 Linear latent variate models . 27 1 Introduction In this paper we discuss statistical inference of moment structures where first and/or second population moments are hypothesized to have a parametric structure. Classical examples of such models are multinomial and covariance structures models. The presented theory is sufficiently general to handle various situations, however the main focus is on covariance structures. Theory and applications of covariance structures were motivated first by the factor analysis model and its various generalizations and later by a development of the LISREL models (J¨oreskog [15, 16]) (see also Browne [8] for a thorough discussion of covariance structures modeling). -
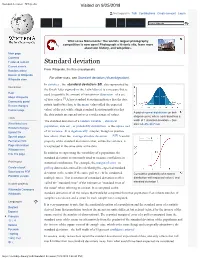
Standard Deviation - Wikipedia Visited on 9/25/2018
Standard deviation - Wikipedia Visited on 9/25/2018 Not logged in Talk Contributions Create account Log in Article Talk Read Edit View history Wiki Loves Monuments: The world's largest photography competition is now open! Photograph a historic site, learn more about our history, and win prizes. Main page Contents Featured content Standard deviation Current events Random article From Wikipedia, the free encyclopedia Donate to Wikipedia For other uses, see Standard deviation (disambiguation). Wikipedia store In statistics , the standard deviation (SD, also represented by Interaction the Greek letter sigma σ or the Latin letter s) is a measure that is Help used to quantify the amount of variation or dispersion of a set About Wikipedia of data values.[1] A low standard deviation indicates that the data Community portal Recent changes points tend to be close to the mean (also called the expected Contact page value) of the set, while a high standard deviation indicates that A plot of normal distribution (or bell- the data points are spread out over a wider range of values. shaped curve) where each band has a Tools The standard deviation of a random variable , statistical width of 1 standard deviation – See What links here also: 68–95–99.7 rule population, data set , or probability distribution is the square root Related changes Upload file of its variance . It is algebraically simpler, though in practice [2][3] Special pages less robust , than the average absolute deviation . A useful Permanent link property of the standard deviation is that, unlike the variance, it Page information is expressed in the same units as the data. -

Sampling Designs for National Forest Assessments Ronald E
Knowledge reference for national forest assessments Sampling designs for national forest assessments Ronald E. McRoberts,1 Erkki O. Tomppo,2 and Raymond L. Czaplewski 3 THIS CHAPTER DISCUSSES THE FOLLOWING POINTS: • Definition of a population for sampling purposes. • Selection of the size and shape for a plot configuration. • Distinguishing among simple random, systematic, stratified and cluster sampling designs. • Methods for constructing sampling design. • Estimating population means and variances. • Estimating sampling errors. • Special considerations for tropical forest inventories. Abstract estimation methods is a key component of the overall process for information management National forest assessments are best conducted and data registration for NFAs (see chapter with sufficiently accurate and scientifically on Information management and data defensible estimates of forest attributes. This registration, p. 93). chapter discusses the statistical design of the sampling plan for a forest inventory, including 1.1 Objectives the process used to define the population to be The goal is to estimate the condition of forests sampled and the selection of a sample intended for an entire nation using data collected from to satisfy NFA precision requirements. The a sample of field plots. The basic objectives team designing a national forest inventory of an NFA are assumed to be fourfold: (i) to should include an experienced statistician. If obtain national estimates of the total area such an expert is not available, this section of forest, subdivided by major categories provides guidance and recommendations for of forest types and conditions, as well as relatively simple sampling designs that reduce the numbers and distributions of trees by risk and improve chances for success. -

Calculating 95% Upper Confidence Limit (UCL) Guidance Document
STATE OF CONNECTICUT DEPARTMENT OF ENERGY AND ENVIRONMENTAL PROTECTION Guidance for Calculating the 95% Upper Confidence Level for Demonstrating Compliance with the Remediation Standard Regulations May 30, 2014 Robert J. Klee, Commissioner 79 Elm Street, Hartford, CT 06106 www.ct.gov/deep/remediation 860-424-3705 TABLE OF CONTENTS LIST OF ACRONYMS iii DEFINITION OF TERMS iv 1. Introduction 1 1.1 Definition of 95 % Upper Confidence Level 1 1.2 Data Quality Considerations 2 1.3 Applicability 2 1.3.1 Soil 3 1.3.2 Groundwater 3 1.4 Document Organization 4 2. Developing a Data Set for a Release Area 4 2.1 Data Selection for 95% UCL Calculation for a Release Area 5 2.2 Non-Detect Soil Results in a Release Area Data Set 7 2.3 Quality Control Soil Sample Results in a Release Area Data Set 7 3. Developing a Data Set for a Groundwater Plume 7 3.1 Data Selection for 95% UCL Calculation for a Groundwater Plume 8 3.2 Non-Detect Results in a Groundwater Plume Data Set 8 3.3 Quality Control Results in a Groundwater Plume Data Set 8 4. Evaluating the Data Set 9 4.1 Distribution of COC Concentrations in the Environment 9 4.2 Appropriate Data Set Size 10 4.3 Statistical DQOs 10 4.3.1 Randomness of Data Set 10 4.3.2 Strength of Data Set 10 4.3.3 Skewness of Data Set 11 5. Statistical Calculation Methods 11 i 5.1 Data Distributions 12 5.2 Handling of Non-Detect Results in Statistical Calculations 12 6. -

Introduction to Estimation
5: Introduction to Estimation Parameters and statistics Statistical inference is the act of generalizing from a sample to a population with calculated degree of certainty. The two forms of statistical inference are estimation and hypothesis testing. This chapter introduces estimation. The next chapter introduces hypothesis testing. A statistical population represents the set of all possible values for a variable. In practice, we do not study the entire population. Instead, we use data in a sample to shed light on the wider population. The process of generalizing from the sample to the population is statistical inference. The term parameter is used to refer to a numerical characteristic of a population. Examples of parameters include the population mean (μ) and the population standard deviation (σ). A numerical characteristic of the sample is a statistic. In this chapter we introduce a particular type of statistic called an estimate. The sample mean x is the natural estimator of population mean μ. Sample standard deviation s is the natural estimator of population standard deviation σ. Different symbols are used to denote parameters and estimates. (e.g., μ versus x ). The parameter is a fixed constant. In contrast, the estimator varies from sample to sample. Other differences are summarized: Parameters Estimators Source Population Sample Value known? No Yes (calculate) Notation Greek (μ) Roman ( x ) Vary from sample to sample No Yes Error-prone No Yes Page 5.1 (C:\data\StatPrimer\estimation.doc, 8/1/2006) Sampling distribution of a mean (SDM) If we had the opportunity to take repeated samples from the same population, samples means ( x s) would vary from sample to sample and form a sampling distribution means (SDM). -

Exercise 1C Scientific Investigation: Statistical Analysis
Exercise 1C Scientific Investigation: Statistical Analysis Parts of this lab adapted from General Ecology Labs, Dr. Chris Brown, Tennessee Technological University and Ecology on Campus, Dr. Robert Kingsolver, Bellarmine University. In part C of our Scientific Investigation labs, we will use the measurement data from part B to ask new questions and apply some basic statistics. Ecology is the ambitious attempt to understand life on a grand scale. We know that the mechanics of the living world are too vast to see from a single vantage point, too gradual to observe in a single lifetime, and too complex to capture in a single narrative. This is why ecology has always been a quantitative discipline. Measurement empowers ecologists because our measuring instruments extend our senses, and numerical records extend our capacity for observation. With measurement data, we can compare the growth rates of trees across a continent, through a series of environmental conditions, or over a period of years. Imagine trying to compare from memory the water clarity of two streams located on different continents visited in separate years, and you can easily appreciate the value of measurement. Numerical data extend our capacity for judgment too. Since a stream runs muddier after a rain and clearer in periods of drought, how could you possibly wade into two streams in different seasons and hope to develop a valid comparison? In a world characterized by change, data sets provide reliability unrealized by single observations. Quantitative concepts such as averages, ratios, variances, and probabilities reveal ecological patterns that would otherwise remain unseen and unknowable. -

Quality Guidelines for Frames Insocialstatistics
Quality Guidelines for Frames in Social Statistics Version 1.51 ESSnet KOMUSO Quality in Multisource Statistics http://ec.europa.eu/eurostat/cros/content/essnet-quality-multisource-statistics_en Framework Partnership Agreement No 07112.2015.003-2015.226 Specific Grant Agreement No 3 (SGA-3) No 07112.2018.007-2018.0444 Work Package 2 QUALITY GUIDELINES FOR FRAMES IN SOCIAL STATISTICS Version 1.51, 2019-09-30 Prepared by: Thomas Burg (Statistics Austria), Alexander Kowarik (Statistics Austria), Magdalena Six (Statistics Austria), Giovanna Brancato (Istat, Italy) and Danuté Krapavickaité (LS, Lithuania) Reviewers: Fabian Bach (ESTAT), Lionel Vigino (ESTAT), Li-Chun Zhang (SSB, Norway), Ton de Waal (CBS, The Netherlands), Angela McCourt (CSO, Ireland), Eva-Maria Asamer (Statistics Austria) and Christoph Waldner (Statistics Austria) ESSnet co-ordinator: Niels Ploug (DST, Denmark), email [email protected], telephone +45 3917 3951 1 Quality Guidelines for Frames in Social Statistics Version 1.51 Table of contents 1 Purpose and Objectives of the Document ................................................................................................................ 3 2 Frames in Official Statistics ....................................................................................................................................... 5 2.1 Definition of a Frame ........................................................................................................................................ 5 2.2 Frames in Social Statistics ................................................................................................................................ -

Is the 'N = 30 Rule of Thumb' of Ecological Field Studies Reliable? a Call for Greater Attention to the Variability in Our Data
Animal Biodiversity and Conservation 37.1 (2014) Forum95 Is the 'n = 30 rule of thumb' of ecological field studies reliable? A call for greater attention to the variability in our data A. Martínez–Abraín Martínez–Abraín, A., 2014. Is the 'n = 30 rule of thumb' of ecological field studies reliable? A call for greater attention to the variability in our data. Animal Biodiversity and Conservation, 37.1: 95–100, Doi: https://doi. org/10.32800/abc.2014.37.0095 Abstract Is the 'n = 30 rule of thumb' of ecological field studies reliable? A call for greater attention to the variability in our data.— A common practice of experimental design in field ecology, which relies on the Central Limit Theorem, is the use of the 'n = 30 rule of thumb'. I show here that papers published in Animal Biodiversity and Conservation during the period 2010–2013 adjust to this rule. Samples collected around this relatively small size have the advantage of coupling statistically–significant results with large effect sizes, which is positive because field researchers are commonly interested in large ecological effects. However, the power to detect a large effect size depends not only on sample size but, importantly, also on between–population variability. By means of a hypothetical example, I show here that the statistical power is little affected by small–medium variance changes between populations. However, power decreases abruptly beyond a certain threshold, which I identify roughly around a five–fold difference in variance between populations. Hence, researchers should explore variance profiles of their study populations to make sure beforehand that their study populations lies within the safe zone to use the 'n = 30 rule of thumb'.