1 Telomeres in Evolution and Development From
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of 3D Genomic Interactions Identifies Candidate Host Genes That Transposable Elements Potentially Regulate Ramya Raviram1,2,3†, Pedro P
Raviram et al. Genome Biology (2018) 19:216 https://doi.org/10.1186/s13059-018-1598-7 RESEARCH Open Access Analysis of 3D genomic interactions identifies candidate host genes that transposable elements potentially regulate Ramya Raviram1,2,3†, Pedro P. Rocha1,4†, Vincent M. Luo1,2, Emily Swanzey5, Emily R. Miraldi2,6,7,8, Edward B. Chuong9, Cédric Feschotte10, Richard Bonneau2,6,7 and Jane A. Skok1* Abstract Background: The organization of chromatin in the nucleus plays an essential role in gene regulation. About half of the mammalian genome comprises transposable elements. Given their repetitive nature, reads associated with these elements are generally discarded or randomly distributed among elements of the same type in genome-wide analyses. Thus, it is challenging to identify the activities and properties of individual transposons. As a result, we only have a partial understanding of how transposons contribute to chromatin folding and how they impact gene regulation. Results: Using PCR and Capture-based chromosome conformation capture (3C) approaches, collectively called 4Tran, we take advantage of the repetitive nature of transposons to capture interactions from multiple copies of endogenous retrovirus (ERVs) in the human and mouse genomes. With 4Tran-PCR, reads are selectively mapped to unique regions in the genome. This enables the identification of transposable element interaction profiles for individual ERV families and integration events specific to particular genomes. With this approach, we demonstrate that transposons engage in long-range intra-chromosomal interactions guided by the separation of chromosomes into A and B compartments as well as topologically associated domains (TADs). In contrast to 4Tran-PCR, Capture-4Tran can uniquely identify both ends of an interaction that involve retroviral repeat sequences, providing a powerful tool for uncovering the individual transposable element insertions that interact with and potentially regulate target genes. -

Reverse Transcriptase Activity Innate to DNA Polymerase I and DNA
Reverse transcriptase activity innate to DNA SEE COMMENTARY polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex Kai Bao*† and Stanley N. Cohen*‡§ Departments of *Genetics and ‡Medicine, Stanford University School of Medicine, Stanford, CA 94305-5120 Edited by Nicholas R. Cozzarelli, University of California, Berkeley, CA, and approved August 10, 2004 (received for review June 18, 2004) Replication of Streptomyces linear chromosomes and plasmids gation and, remarkably, that both of these eubacterial enzymes -proceeds bidirectionally from a central origin, leaving recessed 5 can function efficiently as RTs in addition to having the bio termini that are extended by a telomere binding complex. This chemical properties predicted from their sequences. We further complex contains both a telomere-protecting terminal protein show that the Streptomyces coelicolor and Streptomyces lividans (Tpg) and a telomere-associated protein that interacts with Tpg TopA proteins are prototypes for a subfamily of bacterial and the DNA ends of linear Streptomyces replicons. By using topoisomerases whose catalytic domains contain a unique Asp– histidine-tagged telomere-associated protein (Tap) as a scaffold, Asp doublet motif that is required for their RT activity and which we identified DNA polymerase (PolA) and topoisomerase I (TopA) is essential also to the RNA-dependent DNA polymerase func- proteins as other components of the Streptomyces telomere com- tions of HIV RT and eukaryotic cell telomerases (16, 17). plex. Biochemical characterization of these proteins indicated that both PolA and TopA exhibit highly efficient reverse transcriptase Materials and Methods (RT) activity in addition to their predicted functions. Although RT Plasmid and Bacterial Strains. -

Development of Adenoviral Vectors for Monitoring Telomerase Activity in Living Cells
Development of adenoviral vectors for monitoring telomerase activity in living cells Edqvist, Anna 2007 Link to publication Citation for published version (APA): Edqvist, A. (2007). Development of adenoviral vectors for monitoring telomerase activity in living cells. Genetics. Total number of authors: 1 General rights Unless other specific re-use rights are stated the following general rights apply: Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal Read more about Creative commons licenses: https://creativecommons.org/licenses/ Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. LUND UNIVERSITY PO Box 117 221 00 Lund +46 46-222 00 00 Download date: 05. Oct. 2021 DEVELOPMENT OF ADENOVIRAL VECTORS FOR MONITORING TELOMERASE ACTIVITY IN LIVING CELLS ANNA HELENA EDQVIST 2007 DEPARTMENT OF CELL AND ORGANISM BIOLOGY LUND UNIVERSITY SWEDEN A doctoral thesis at a university in Sweden is produced either as a monograph or as a collection of papers. In the latter case, the introductory part constitutes the formal thesis, which summarises the accompanying papers. -

Microorganisms
microorganisms Review Rules and Exceptions: The Role of Chromosomal ParB in DNA Segregation and Other Cellular Processes Adam Kawalek y , Pawel Wawrzyniak y, Aneta Agnieszka Bartosik and Grazyna Jagura-Burdzy * Department of Microbial Biochemistry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawi´nskiego5a, 02-106 Warsaw, Poland; [email protected] (A.K.); [email protected] (P.W.); [email protected] (A.A.B.) * Correspondence: [email protected]; Tel.: +48-225921212 These authors contributed equally to this work. y Received: 4 December 2019; Accepted: 9 January 2020; Published: 11 January 2020 Abstract: The segregation of newly replicated chromosomes in bacterial cells is a highly coordinated spatiotemporal process. In the majority of bacterial species, a tripartite ParAB-parS system, composed of an ATPase (ParA), a DNA-binding protein (ParB), and its target(s) parS sequence(s), facilitates the initial steps of chromosome partitioning. ParB nucleates around parS(s) located in the vicinity of newly replicated oriCs to form large nucleoprotein complexes, which are subsequently relocated by ParA to distal cellular compartments. In this review, we describe the role of ParB in various processes within bacterial cells, pointing out interspecies differences. We outline recent progress in understanding the ParB nucleoprotein complex formation and its role in DNA segregation, including ori positioning and anchoring, DNA condensation, and loading of the structural maintenance of chromosome (SMC) proteins. The auxiliary roles of ParBs in the control of chromosome replication initiation and cell division, as well as the regulation of gene expression, are discussed. Moreover, we catalog ParB interacting proteins. -

Telomere-Loop Dynamics in Chromosome End Protection
bioRxiv preprint doi: https://doi.org/10.1101/279877; this version posted March 9, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Telomere-loop dynamics in chromosome end protection David Van Ly1,2, Ronnie Ren Jie Low1,6, Sonja Frölich1,7, Tara K. Bartolec1,8, Georgia R. Kafer1, Hilda A. Pickett3, Katharina Gaus4,5, and Anthony J. Cesare1,9*. 1Genome Integrity Unit, Children’s Medical Research Institute, University of Sydney, Westmead, New South Wales 2145, Australia. 2 School of Medicine, The University of Notre Dame Australia, Sydney, New South Wales 2010, Australia. 3Telomere Length Regulation Unit, Children’s Medical Research Institute, University of Sydney, Westmead, New South Wales 2145, Australia. 4EMBL Australia Node in Single Molecule Science, School of Medical Sciences, University of New South Wales, Sydney, NSW 2052, Australia. 5ARC Centre of Excellence in Advanced Molecular Imaging, University of New South Wales, Sydney, NSW 2052, Australia. 6Current affiliation: The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria 3052, Australia 7Current affiliation: Robinson Research Institute, School of Medicine, University of Adelaide, Adelaide, South Australia 5005, Australia. 8Current affiliation: School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, New South Wales, 2052, Australia 9Lead contact *Correspondence: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/279877; this version posted March 9, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

The NIH Catalyst from the Deputy Director for Intramural Research
— Fostering Communication and Collaboration The nihCatalyst A Publication fob NIH Intramural Scientists National Institutes of Health i Office of the Director Volume 8, Issue 6 November-December 2000 NIH Research Festival Translational Research Running with the Genome: Jean Li d Cadet: NIH Gets To Work with the ‘Working Draft’ A Bridge at NIDA by Cynthia Delgado by Fran Pollner he “working draft” se- r ean Lud Cadet is equally at home quence of the human I upstairs and downstairs in the genome, delivered to building that houses the NIDA in- T | the desktops of the biomedi- Jamural research program at the cal community this summer, Johns Hopkins University Bayview may become the most “dog- campus in Baltimore. eared” work-in-progress the On the fourth floor, Cadet runs the world has ever known. four labs that make up the Molecu- Unlike the first draft of a lar Neuropsychiatry Unit, of which medical text that requires cor- he is chief; and on the first floor, he rections and updates before directs NIDA’s clinical research pro- it is finally printed, the gram, overseeing the inpatient re- genome’s first draft is out search unit, the outpatient research there, and some NIH investi- program, an adolescent clinic that fo- gators are using it to make cuses on smoking new discoveries while oth- cessation, and the — ers are arranging the data to 70 currently active make the material even more clinical protocols meaningful as a resource to all of which are the research community. strategic use of the HGP’s data aided conducted on-site. -

Robust Measurement of Telomere Length in Single Cells PNAS PLUS
Robust measurement of telomere length in single cells PNAS PLUS Fang Wanga,b, Xinghua Panc,1, Keri Kalmbacha, Michelle L. Seth-Smitha, Xiaoying Yeb, Danielle M. F. Antumesa,d,e, Yu Yinb, Lin Liub,1, David L. Keefea,1, and Sherman M. Weissmanc,1 aDepartment of Obstetrics and Gynecology, New York University Langone Medical Center, NY 10016; bState Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin 300071, China; cDepartment of Genetics, Yale University School of Medicine, New Haven, CT 06520-8005; dDepartment of Pathology, Fluminense Federal University, 24033-900, Rio de Janeiro, Brazil; and eCoordenacao de Aperfeicoamento de Pessoal de Nivel Superior (CAPES) Foundation, Ministry of Education of Brazil, 70040-020, Brasilia, Brazil Contributed by Sherman M. Weissman, April 9, 2013 (sent for review February 5, 2013) Measurement of telomere length currently requires a large popula- length for a given cell population (10–12). High-throughput tion of cells, which masks telomere length heterogeneity in single Q-FISH, flow FISH, and single telomere length analysis can be used cells, or requires FISH in metaphase arrested cells, posing technical for telomere measurement of dividing, nondividing, and senescent challenges. A practical method for measuring telomere length in cells, but these methods also require large cell populations (13–15). single cells has been lacking. We established a simple and robust The ability to measure telomere length in single cells rather approach for single-cell telomere length measurement (SCT-pqPCR). than relying upon average telomere length in cell populations or We first optimized a multiplex preamplification specific for telo- the entire tissue enables the study of biological heterogeneity on meres and reference genes from individual cells, such that the ampli- a cell-by-cell basis, an issue of fundamental importance for studies con provides a consistent ratio (T/R) of telomeres (T) to the reference of aging, development, carcinogenesis, and many other diseases. -

DNA Viral Genomes • As Small As 1300 Nt in Length Ssrna Viral Genomes • Smallest Sequenced Genome = 2300 Nt • Largest = 31,000 Nt
Viral Genome Organization All biological organisms have a genome • The genome can be either DNA or RNA • Encode functions necessary o to complete its life cycle o interact with the environments Variation • Common feature when comparing genomes Genome sequencing projects • Uncovering many unique features • These were previously known. General Features of Viruses Over 4000 viruses have been described • Classified into 71 taxa • Some are smaller than a ribosome Smallest genomes • But exhibit great variation Major classifications • DNA vs RNA Sub classifications • Single-stranded vs. double-stranded Segments • Monopartitie vs. multipartite ssRNA virus classifications • RNA strand found in the viron o positive (+) strand . most multipartite o negative (-) strand . many are multipartite Virus replication by • DNA viruses o DNA polymerase . Small genomes • Host encoded . Large genomes • Viral encoded • RNA viruses o RNA-dependent RNA polymerase o Reverse transcriptase (retroviruses) Genome sizes • DNA viruses larger than RNA viruses ss viruses smaller than ds viruses • Hypothesis • ss nucleic acids more fragile than ds nucleic • drove evolution toward smaller ss genomes Examples: ssDNA viral genomes • As small as 1300 nt in length ssRNA viral genomes • Smallest sequenced genome = 2300 nt • Largest = 31,000 nt Mutations • RNA is more susceptible to mutation during transcription than DNA during replication • Another driving force to small RNA viral genomes Largest genome • DNA virus • Paramecium bursaria Chlorella virus 1 o dsDNA o 305,107 nt o 698 proteins Table 1. General features of sequenced viral genomes. The data was collected from NCBI (http://www.ncbi.nlm.nih.gov/genomes/VIRUSES/viruses.html) and represents the October 13, 2014 release from NCBI. -

Cshperspect-MEV-A018168 1..17
Downloaded from http://cshperspectives.cshlp.org/ on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press Coevolution of the Organization and Structure of Prokaryotic Genomes Marie Touchon1,2 and Eduardo P.C. Rocha1,2 1Microbial Evolutionary Genomics, Institut Pasteur, 75015 Paris, France 2CNRS, UMR3525, 75015 Paris, France Correspondence: [email protected] The cytoplasm of prokaryotes contains many molecular machines interacting directly with the chromosome. These vital interactions depend on the chromosome structure, as a mole- cule, and on the genome organization, as a unit of genetic information. Strong selection for the organization of the genetic elements implicated in these interactions drives replicon ploidy, gene distribution, operon conservation, and the formation of replication-associated traits. The genomes of prokaryotes are also very plastic with high rates of horizontal gene transfer and gene loss. The evolutionary conflicts between plasticity and organization lead to the formation of regions with high genetic diversity whose impact on chromosome structure is poorly understood. Prokaryotic genomes are remarkable documents of natural history because they carry the imprint of all of these selective and mutational forces. Their study allows a better understanding of molecular mechanisms, their impact on microbial evolu- tion, and how they can be tinkered in synthetic biology. rokaryotic cells typically lack a clear physi- nus in the cell. As replication proceeds, DNA Pcal separation between DNA and the cyto- regions are under different states of replication plasm. The cell is therefore a complex network and are being segregated in function of the of genetic and biochemical interactions involv- growing multiple division septa. Hence, repli- ing the DNA molecules and many cellular pro- cation, segregation, and cell doubling are tightly cesses. -

The Evolution of the Mitochondrial Genomes of Calcareous Sponges and Cnidarians Ehsan Kayal Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2012 The evolution of the mitochondrial genomes of calcareous sponges and cnidarians Ehsan Kayal Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Evolution Commons, and the Molecular Biology Commons Recommended Citation Kayal, Ehsan, "The ve olution of the mitochondrial genomes of calcareous sponges and cnidarians" (2012). Graduate Theses and Dissertations. 12621. https://lib.dr.iastate.edu/etd/12621 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. The evolution of the mitochondrial genomes of calcareous sponges and cnidarians by Ehsan Kayal A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Ecology and Evolutionary Biology Program of Study Committee Dennis V. Lavrov, Major Professor Anne Bronikowski John Downing Eric Henderson Stephan Q. Schneider Jeanne M. Serb Iowa State University Ames, Iowa 2012 Copyright 2012, Ehsan Kayal ii TABLE OF CONTENTS ABSTRACT .......................................................................................................................................... -
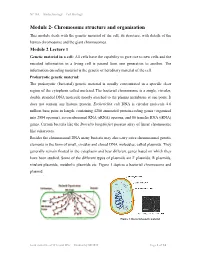
Chromosome Structure and Organisation
NPTEL – Biotechnology – Cell Biology Module 2- Chromosome structure and organisation This module deals with the genetic material of the cell, its structure, with details of the human chromosome and the giant chromosomes. Module 2 Lecture 1 Genetic material in a cell: All cells have the capability to give rise to new cells and the encoded information in a living cell is passed from one generation to another. The information encoding material is the genetic or hereditary material of the cell. Prokaryotic genetic material: The prokaryotic (bacterial) genetic material is usually concentrated in a specific clear region of the cytoplasm called nucleiod. The bacterial chromosome is a single, circular, double stranded DNA molecule mostly attached to the plasma membrane at one point. It does not contain any histone protein. Escherichia coli DNA is circular molecule 4.6 million base pairs in length, containing 4288 annotated protein-coding genes (organized into 2584 operons), seven ribosomal RNA (rRNA) operons, and 86 transfer RNA (tRNA) genes. Certain bacteria like the Borrelia burgdorferi possess array of linear chromosome like eukaryotes. Besides the chromosomal DNA many bacteria may also carry extra chromosomal genetic elements in the form of small, circular and closed DNA molecules, called plasmids. They generally remain floated in the cytoplasm and bear different genes based on which they have been studied. Some of the different types of plasmids are F plasmids, R plasmids, virulent plasmids, metabolic plasmids etc. Figure 1 depicts a bacterial chromosome and plasmid. Figure 1: Bacterial genetic material Joint initiative of IITs and IISc – Funded by MHRD Page 1 of 24 NPTEL – Biotechnology – Cell Biology Virus genetic material: The chromosomal material of viruses is DNA or RNA which adopts different structures. -

Mitochondrial Genome of Non-Photosynthetic Mycoheterotrophic Plant Hypopitys 1 Monotropa,Its Structure, Gene Expression And
bioRxiv preprint doi: https://doi.org/10.1101/681718; this version posted December 3, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Mitochondrial genome of non-photosynthetic mycoheterotrophic plant Hypopitys 2 monotropa,its structure, gene expression and RNA editing 3 4 Viktoria Y. Shtratnikova1, Mikhail I. Schelkunov2,3, Aleksey A. Penin2, Maria D. Logacheva1,2,3 5 6 1A. N. Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 7 Moscow, Russian Federation 8 2Laboratory of Plant Genomics, Institute for Information Transmission Problems of the Russian 9 Academy of Sciences, Moscow, Russian Federation 10 3Skolkovo Institute of Science and Technology, Moscow, Russian Federation 11 12 Corresponding Author: 13 Maria D. Logacheva1,2,3 14 1Lomonosov Moscow State University, Leninskie Gory, GSP-1, Moscow 119991, Russia 15 2BolshoyKaretny lane, 19/1, Moscow 127051, Russia 16 3Skolkovo Institute of Science and Technology, Nobel St. 3, Moscow 143026, Russia 17 18 Email address: [email protected] 19 20 Abstract 21 Heterotrophic plants – the plants that lost the ability to photosynthesis – are characterized by a 22 number of changes at all levels of organization. Heterotrophic plants divide into two large 23 categories – parasitic and mycoheterotrophic. The question of to what extent these changes are 24 similar in these two categories is still open. Plastid genomes of non-photosynthetic plants are 25 well characterized and they demonstrate similar patterns of reduction in both groups. In contrast, 26 little is known about mitochondrial genomes of mycoheterotrophic plants.