Investigating and Assessing the Dermoepidermal Junction with Multiphoton Microscopy and Deep Learning
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Skin Chapter Goals
2/4/2016 Chapter 16: Skin Find this out on page 650 in your book: What the name for the system that includes skin? How much does our skin weigh? How much surface area does it cover? Copyright © 2011, 2008, 2005 by Saunders, an imprint of Elsevier Inc. All rights reserved. 1 Chapter Goals Name the layers of the skin and the accessory structures associated with the skin. Build medical words using the combining forms that are related to the specialty of dermatology. Identify lesions, signs, and symptoms, and pathologic conditions that relate to the skin. Copyright © 2011, 2008, 2005 by Saunders, an imprint of Elsevier Inc. All rights reserved. 2 1 2/4/2016 Chapter Goals Describe laboratory tests and clinical procedures that pertain to the skin and recognize relevant abbreviations. Apply your new knowledge to understanding medical terms in their proper contexts, such as medical reports and records. Copyright © 2011, 2008, 2005 by Saunders, an imprint of Elsevier Inc. All rights reserved. 3 Introduction ● the skin and its accessory structures (hair, nails and glands) make up the integumentary system of the body ● weighs 8-10 lb ● covers 22 square feet Copyright © 2011, 2008, 2005 by Saunders, an imprint of Elsevier Inc. All rights reserved. 4 2 2/4/2016 Functions of Skin provides protective membrane - guards the deeper tissues against excessive loss of water, salts and heat - protects against pathogens glands lubricate and cool the skin receptor for sensations (pain, temp, pressure and touch) helps maintain body temperature (thermoregulation) Copyright © 2011, 2008, 2005 by Saunders, an imprint of Elsevier Inc. -

Anatomy and Physiology of Hair
Chapter 2 Provisional chapter Anatomy and Physiology of Hair Anatomy and Physiology of Hair Bilgen Erdoğan ğ AdditionalBilgen Erdo informationan is available at the end of the chapter Additional information is available at the end of the chapter http://dx.doi.org/10.5772/67269 Abstract Hair is one of the characteristic features of mammals and has various functions such as protection against external factors; producing sebum, apocrine sweat and pheromones; impact on social and sexual interactions; thermoregulation and being a resource for stem cells. Hair is a derivative of the epidermis and consists of two distinct parts: the follicle and the hair shaft. The follicle is the essential unit for the generation of hair. The hair shaft consists of a cortex and cuticle cells, and a medulla for some types of hairs. Hair follicle has a continuous growth and rest sequence named hair cycle. The duration of growth and rest cycles is coordinated by many endocrine, vascular and neural stimuli and depends not only on localization of the hair but also on various factors, like age and nutritional habits. Distinctive anatomy and physiology of hair follicle are presented in this chapter. Extensive knowledge on anatomical and physiological aspects of hair can contribute to understand and heal different hair disorders. Keywords: hair, follicle, anatomy, physiology, shaft 1. Introduction The hair follicle is one of the characteristic features of mammals serves as a unique miniorgan (Figure 1). In humans, hair has various functions such as protection against external factors, sebum, apocrine sweat and pheromones production and thermoregulation. The hair also plays important roles for the individual’s social and sexual interaction [1, 2]. -

Important Functions of the Skin Why Dermatology?
Introduction to Learning objectives • To Understand the basic structure of the skin Dermatology • To acquire the basic knowledge of the skin functions • Be able to describe skin lesions and presentations properly • Be familiar with the standard diagnostic tools in dermatology Dr. Sami N. Alsuwaidan ASSCOCIATE PROFESSOR AND CONSULTANT IN DERMATOLOGY AND LASER SURGERY DEPARTMENT OF DERMATOLOGY KING SAUD UNIVERSITY RIYADH, SAUDIA ARABIA 2 Important functions of the Dermatology skin Skin, hair, nails, and mucous - Protection against external injury membranes (mouth and genitila). - Fluid balance - Temperature buffering - Synthesis of Vit. D - Immune system - Cosmetic function 3 4 Cornified layer Epidermis Granular layer Why Dermatology ? Spinous layer Dermis Basal layer 5 6 1 1 Skin Anatomy 1 Epidermis 2 Basement membrane (dermoepidermal junction) Epidermis 3 Dermis 4 Subcutaneous fat Epidermis: Four layers (from outside – inside) 1. Cornified layer 2. Granular layer Dermis 3. Spinous layer 4. Basal layer Dermis contains: 1. Collagen fibers 2. Elastic fibers 3. Ground substances 4. Blood vessels 5. Nerves. Subcutaneous 7 8 Skin Appendages Hair follicle Sebaceous gland Arrector Pilli muscle Arrector pili muscle Eccrine sweat gland Hair follicle Apocrine sweat glands 9 10 Nail Anatomy Sebaceous gland Eccrine gland Apocrine gland 11 12 1 2 Examination Primary Lesions 1. Morphology 2. Configuration Secondary lesions 3. Distribution 13 14 Primary Lesions Macule Papule Plaque Nodule Wheal Vesicle Bulla Pustule 15 16 a 17 a 1 3 19 20 a a 21 22 23 24 1 4 Secondary lesions Crust Scale Ulceration Excoriation Scar Fissure Lichenification 25 26 28 a 30 a 1 5 31 32 34 35 1 6 38 Color and Shape Distribution Configuration 39 40 41 42 1 7 43 44 45 46 Dermatographism : When you stroke the Some specific signs in normal skin edema and erythema (you can write on skin!) .Seen in physical urticaria Dermatology Kobener Phenomenon : Induction of new skin lesions on previously normal appearing skin by truma e.g. -

Histology of Juvenile Skin of Lepidosiren Paradoxa Fitzinger, 1837 (Sarcopterygii, Dipnoi)
Anais da Academia Brasileira de Ciências (2019) 91(4): e20190822 (Annals of the Brazilian Academy of Sciences) Printed version ISSN 0001-3765 / Online version ISSN 1678-2690 http://dx.doi.org/10.1590/0001-3765201920190822 www.scielo.br/aabc | www.fb.com/aabcjournal Histology of juvenile skin of Lepidosiren paradoxa Fitzinger, 1837 (Sarcopterygii, Dipnoi) LUIS ALBERTO ROMANO1, ANDREA I.H. LÓPEZ1, JUAN RAFAEL BUITRAGO2 and VIRGÍNIA F. PEDROSA1 1Institute of the Oceanography, University Federal of the Rio Grande, Laboratory of the de Immunology and Pathology of the Aquatic Organisms, Rua do Hotel, 2, Cassino, 96210-030 Rio Grande, RS, Brazil 2University Federal of the Rio Grande, Laboratory of the Biochemistry Functional of Aquatic Organisms, Rua do Hotel, 2, Cassino, 96210-030 Rio Grande, RS, Brazil Manuscript received on July 20, 2019; accepted for publication on September 24, 2019 How to cite: ROMANO LA, LÓPEZ AIH, BUITRAGO JR AND PEDROSA VF. 2019. Histology of juvenile skin of Lepidosiren paradoxa Fitzinger, 1837 (Sarcopterygii, Dipnoi). An Acad Bras Cienc 91: e20190822. DOI 10.1590/0001-3765201920190822. Abstract: The skin of three juvenile Lepidosiren paradoxa specimens was examined. The epidermis was composed of a polystratified epithelium resting on a basement membrane, including mucus-secreting cells, and a cuticle of mucopolysaccharides on the surface. Two types of skin receptors, electroreceptors and mechanoreceptors, were found; the first type was located in the dermoepidermal junction, and the second type was completely intraepiderma. The skin structure of these fish, suggests the possibility of the skin participating in the breath. Key words: electroreceptors, lungfish, mechanoreceptors, Paraná River basin, pirambóia. -

Clinical and Onychoscopic Features of Benign and Malignant Conditions in Longitudinal Melanonychia in the Thai Population: a Comparative Analysis
Clinical, Cosmetic and Investigational Dermatology Dovepress open access to scientific and medical research Open Access Full Text Article ORIGINAL RESEARCH Clinical and Onychoscopic Features of Benign and Malignant Conditions in Longitudinal Melanonychia in the Thai Population: A Comparative Analysis This article was published in the following Dove Press journal: Clinical, Cosmetic and Investigational Dermatology Pintusorn Kungvalpivat1 Background: Longitudinal melanonychia can arise from many underlying conditions, both Salinee Rojhirunsakool1,2 benign and malignant. Practitioners tend to be reluctant to perform a biopsy of this condition due Pamela Chayavichitsilp1 to procedure-related pain and the possibility of permanent nail dystrophy. Onychoscopy has Poonkiat Suchonwanit 1 become a useful tool to provide a provisional diagnosis and assist in deciding on a nail biopsy. Chanitwan T Wichayachakorn1 Objective: To investigate and differentiate the clinical and onychoscopic features of sub ungual melanoma (SUM)/subungual melanoma in situ (SMIS) and other benign melanocytic Suthinee Rutnin 1 conditions (BM). 1 Division of Dermatology, Department of Materials and Methods: In this cross-sectional study, a total of 32 cases of longitudinal Medicine, Faculty of Medicine, Ramathibodi Hospital, Mahidol melanonychia were examined, and baseline characteristics were recorded. Onychoscopic University, Bangkok, Thailand; 2Skin pictures were taken by handheld dermoscopy with 10x and 50x magnification. A biopsy Center, Srinakharinwirot University, was then performed in each case, and a pathological diagnosis was obtained. Bangkok, Thailand Results: Of the 32 cases, 6 were diagnosed with SMIS and 26 with BM (21 simple lentigines, 5 junctional nevi). The median age was significantly higher among the SMIS group (56 vs 31 years) (p = 0.034). -

Hair Follicles
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Modulation of BMP Signaling by Noggin is Required for Induction of the Secondary (Nontylotrich) Hair Follicles Vladimir A. Botchkarev, Natalia V. Botchkareva, Andrei A. Sharov, Keiko Funa,* Otmar Huber,² and Barbara A. Gilchrest Department of Dermatology, Boston University School of Medicine, Boston, Massachusetts, U.S.A.; *Department of Anatomy and Cell Biology, University of Gothenburg, Gothenburg, Sweden; ²Department of Clinical Chemistry and Pathobiochemistry, University Hospital Benjamin Franklin, Free University, Berlin, Germany Increasing evidence suggests that morphogenesis of arrest of primary hair follicle development prior to the distinct developmental structures derived from hair shaft formation. The loss of noggin activity was the same organ-committed epithelium is controlled associated with failure to express genes that specify by differential mechanisms. As was recently shown hair follicle cell fates in the epidermis (Lef-1, b-cate- in mice with mutations in the downless (dL) gene, nin, Shh) and dermal papilla (p75 kDa neurotrophin induction of primary or tylotrich hair follicles is receptor, alkaline phosphatase). This suggests that strikingly dependent of signaling through the Tnf regulation of BMP2/4 signaling by noggin is essential receptor homologue, Edar. Here, we show that dor- for the induction of secondary hair follicles, as well sal skin of murine embryos with constitutive deletion as for advanced stages of development in primary of the BMP2/4 antagonist noggin, after transplanta- hair follicles. Key words: appendage/bone morphogenetic tion into SCID mice, is characterized by the lack of proteins/development/hair follicle/Lef-1/morphogenesis/ induction of secondary hair follicles, and by the Shh/skin/Wnt. -
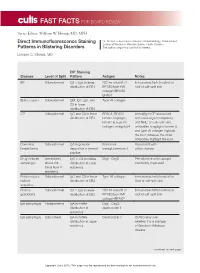
Direct Immunofluorescence Staining Patterns in Blistering Disorders
FAST FACTS FOR BOARD REVIEW Series Editor: William W. Huang, MD, MPH Direct Immunofluorescence Staining Dr. Strowd is Assistant Professor of Dermatology, Wake Forest School of Medicine, Winston-Salem, North Carolina. Patterns in Blistering Disorders The authors report no conflict of interest. Lindsay C. Strowd, MD DIF Staining Disease Level of Split Pattern Antigen Notes BP Subepidermal C3 > IgG in linear NC16a subunit of Immunoreactants located on distribution at DEJ BP180/type XVII roof of salt-split skin collagen/BPAG2, BPAG1 Bullous lupus Subepidermal IgM, IgG, IgA, and Type VII collagen C3 in linear distribution at DEJ CP Subepidermal IgG and C3 in linear BPAG1, BPAG2, Antiepiligrin CP associated distribution at DEJ laminin 5/epiligrin, with solid-organ malignancy laminin 6, type VII and NHL; on salt-split skin, collagen, integrin β4 antibodies to epiligrin/laminin 5 and type VII collagen highlight the floor, whereas the other antibodies highlight the roof Dermatitis Subepidermal IgA in granular Epidermal Associated with herpetiformis deposition in dermal transglutaminase 3 celiac disease papillae Drug-induced Immediately IgG > C3 in netlike Dsg1, Dsg3 Penicillamine and captopril pemphigus above the distribution in lower commonly implicated basal layer in epidermis epidermis Epidermolysis Subepidermal IgG and C3 in linear Type VII collagen Immunoreactants located on bullosa distribution at DEJ floor of salt-split skin acquisita Herpes Subepidermal C3 > IgG in linear NC16a subunit of Immunoreactants located on gestationis distribution at DEJ BP180/type XVII roof of salt-split skin collagen/BPAG2 IgA pemphigus Intraepidermal IgA in netlike Dsg1, Dsg3, distribution in desmocollin 1 epidermis IgA pemphigus Subcorneal IgA in netlike Desmocollin 1 Controversy over distribution in upper whether it is a subtype epidermis of Sneddon-Wilkinson disease continued on next page Copyright Cutis 2016. -

Dermatopathology: Practical and Conceptual | Print View
Dermatopathology: Practical and Conceptual | Print View http://www.derm101.com/dynaweb/resources/milestones/4842/@Generi... Dermatopathology: Practical & Conceptual | January - March 2001 | Volume 7, #1 http://www.derm101.com/ © 2002–2006 Ardor Scribendi, Ltd. All rights reserved. [Press Ctrl-P (PC) or Command-P (Mac) to print] EVOLUTION IN THINKING Criteria for Histopathologic Diagnosis of Melanoma, 1947–2000: A Critique in Historical Perspective Mary Aldrene L. Tan, M.D. A. Bernard Ackerman, M.D. Introduction ". no significant changes in histologic criteria had occurred over time that would explain the rise in melanoma incidence."Rigel DS, Friedman RJ, Kopf AW et al. The incidence of malignant melanoma in the United States: Issues as we approach the 21st century. JAAD 1996;34:839–47. Is the statement of Rigel, Friedman, and Kopf true? A reader should be able to decide on the basis of the evidence that follows. Becker and Obermayer "A primary cutaneous nodule of melanoma is composed of melanoblastic cells which are of two types: the spindle-celled variety (sarcoma-type) as described for lentigo maligna and the round or oval variety (carcinoma-type). Section from an early lesion shows disruption of the epidermodermal junction with increase in number of melanoblastic cells in this region. During the early stage of the process the melanoblastic cells are confined to the superficial portion of the dermis and are limited internally by a moderate to extensive cellular infiltrate composed of lymphocytes and plasma cells. As the tumor enlarges, invasion of the deeper layers of the dermis takes place and the infiltrate decreases in amount. -

Intro/Anatomy of Skin, Hair and Nails
- Stratum spinosum 1 – Intro/Anatomy of o Superficial to stratum basale, named for spiny- appearing desmosomes between cells o Keratins 1 and 10 are expressed in this layer and are skin, hair and nails mutated in epidermolytic hyperkeratosis (aka bullous congenital ichthyosiform erythroderma) Vital Functions - Stratum basale o Located just above the basement membrane, is - Sensation, barrier, immune surveillance, UV protection, composed of 10% stem cells thermoregulation o Keratins 5 and 14 are expressed in the basal layer Fun facts and are mutated in patients with epidermolysis bullosa simplex (EBS) - The skin is the largest human organ, 15% of a person’s body weight Major CELL TYPES of the epidermis - Skin cancer = most common cancer worldwide; affects 1 in 5 1. Keratinocytes people (KC) (“squamous cells”, “epidermal cells”) o Make up most of epidermis, produce keratin - Our skin is constantly being renewed, with the epidermis 2. Melanocytes (MC) turning over q40-56 days, results in average person shedding o Neural-crest derived 9 lbs of skin yearly o Normally present in ratio of 1 MC : 10 KC’s Skin thickness varies based on…. o Synthesize and secrete pigment granules called melanosomes - Location: epidermis is thickest on palms/soles at ~ 1.5mm o *** Different races and skin types actually have the (thickness of a penny), thinnest on eyelid/postauricular at ~ same amount of melanoCYTES but differ in the 0.05mm (paper) number, size, type, and distribution of - Age: Skin is relatively thin in children, thickens up until our melanoSOMES, with fairer skin types having more 30’s or 40’s, and then thins out thereafter. -

Term Topic Content Hair Hair Is One of the Characteristic Features Of
Term Topic content Hair Hair is one of the characteristic features of mammals and has various functions such as protection against external factors; producing sebum, apocrine sweat and pheromones; impact on social and sexual interactions; thermoregulation and being a resource for stem cells. Hair is a derivative of the epidermis and consists of two distinct parts: the follicle and the hair shaft. The follicle is the essential unit for the generation of hair. The hair shaft consists of a cortex and cuticle cells, and a medulla for some types of hairs. Hair follicle has a continuous growth and rest sequence named hair cycle. The duration of growth and rest cycles is coordinated by many endocrine, vascular and neural stimuli and depends not only on localization of the hair but also on various factors, like age and nutritional habits. Distinctive anatomy and physiology of hair follicle are presented in this chapter. Extensive knowledge on anatomical and physiological aspects of hair can contribute to understand and heal different hair disorders. The hair follicle is one of the characteristic features of mammals serves as a unique miniorgan. In humans, hair has various functions such as protection against external factors, sebum, apocrine sweat and pheromones production and thermoregulation. The hair also plays important roles for the individual’s social and sexual interaction. The hair follicle serves as a reservoir for epithelial and melanocyte stem cells and it is capable of being one of the few immune privileged sites of human body. Hair follicle development is related to the interactions between epithelial and mesenchymal cells. -

EFSUMB Course Book, 2Nd Edition
EFSUMB ECB2nd Edition Dermatology... 19.07.2019 12:46 1 EFSUMB Course Book, 2nd Edition Editor: Christoph F. Dietrich Dermatologic Ultrasound Fernando Alfageme1, Ximena Wortsman2, Gastón Roustan1, Maria Crisan3, Radu Badea3 1Dermatology Service. Hospital Universitario Puerta de Hierro Majadahonda. Madrid. Spain 2IDIBAPS. Dermatology and Radiology Department Clinica Seervet. Universidad Católica y Santiago de Chile. Chile 3Radiology and Dermatology UMPh Iuliu Hatieganu, Cluj-Napoca. Romanaia Corresponding author: Prof. Dr. Fernando Alfageme Dermatology Service Hospital Universitario Puerta de Hierro Majadahonda (Madrid) Associate Professor Dermatology Universidad Autónoma de Madrid, Spain EFSUMB ECB2nd Edition Dermatology …. 19.07.2019 12:46 2 Ultrasonography of normal skin and appendages (hair and nails) The skin covers all our human body, but its function goes further than being just a physical barrier. It is necessary to know exactly the dermatologic structures and its ultrasound features. The skin is composed by three anatomically distinct layers: epidermis, dermis and hypodermis or subcutaneous tissue. There are anatomical regional variations of these strata depending on the location (face, trunk, palm or sole). Besides the skin there are the skin appendages, especially the nail and the hair for ultrasound purposes. And finally there are other structures (glands, vessels, nerves) that we will have to take into account in our ultrasound examinations. Maybe the best way to classify skin disease is according to the skin structure affected: epidermis, dermis, subcutaneous fat, hair or nails. We will need a high frequency linear transducer (> 12 MHz) and Doppler for a suitable skin ultrasound study. Epidermis It is the external layer of the skin and it is composed mainly of keratinocytes. -

Effect of Subdermal Pressure on the Skin and Its Appendages in the Sheep
EFFECT OF SUBDERMAL PRESSURE ON THE SKIN AND ITS APPENDAGES IN THE SHEEP By M. JOLLY* and A. G. LYNEt [Manuscript received October 21, 1968] Summary A method was devised by means of which pressure in an outward direction could be exerted on the undersurface of the skin. In each of two sheep (one black, one white) a Perspex disk was inserted subdermally and connected by means of a pin projecting through the skin to a spring supported on a light tower on the animal's back. The device was well tolerated and a pedicle of skin was formed during a period of some months. Macroscopically, there was no change in the skin or wool which showed normal growth, colour, and crimping. No obvious shedding of the fibres occurred. Microscopically, there was a change in fibre criIp.p, particularly in the time taken to form each crimp. There was a decrease of approximately 50 % in the density of the follicle population over the disks but no change in the ratio of secondary to primary follicles. There was no change in length growth rate of fibres over the disks but there was a small increase in the fibre diameters. The epidermis over the disks increased in thickness and a distinct and continuous granular layer formed. Pigmentation of the epidermis was unchanged except near the pin in the black animal, where both melanocytes and melanin were increased. Within the dermis of the black animal, there was an obvious increase in the concentration of melanocytes and melanin adjacent to the pin, both of which were probably derived from degeneration of follicles.