A Comprehensive Molecular Phylogeny for the Hornbills (Aves: Bucerotidae) ⇑ Juan-Carlos T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hornbills of Borneo
The following two species can be easily confused. They can be recognized If you want to support Hornbill Conservation in Sabah, please contact from other hornbill species by the yellow coloration around the head and neck in Marc Ancrenaz at Hutan Kinabatangan Orangutan Conservation Project: the males. The females have black heads and faces and blue throat pouches. [email protected] HORNBILLS OF BORNEO Wrinkled hornbill (Aceros corrugatus): A large, mainly black hornbill whose tail is mostly white with some black at the base. Males have a yellow bill and more prominent reddish casque while females have an all yellow bill and casque. SABAH MALAYSIA The presence of hornbills in the Kinabatangan area is an indication that the surrounding habitat is healthy. Hornbills need forests for nesting and food. Forests need hornbills for dispersal of seeds. And the local people need the forests for wood Wreathed hornbill (Rhyticeros undulatus): A large, primarily black hornbill products, clean water and clean air. They are all connected: whose tail is all white with no black at the base. Both sexes have a pale bill with a small casque and a dark streak/mark on the throat pouch. people, hornbills and forests! Eight different hornbill species occur in Borneo and all are found in Kinabatangan. All are protected from hunting and/or disturbance. By fostering an awareness and concern of their presence in this region, hornbill conservation will be ensured for future generations. Credits: Sabah Forest Department, Sabah Wildlife Department, Hutan Kinabatangan Orangutan Conserva- tion Project (KOCP), Hornbill Research Foundation, Chester Zoo, Woodland Park Zoo. -

Smithsonian Miscellaneous Collections
SMITHSONIAN MISCELLANEOUS COLLECTIONS VOLUME 131, NUMBER 9 BREEDING AND OTHER HABITS OF CASQUED HORNBILLS (BYCANISTES SUBCYLINDRICUS) (With 6 Plates) By LAWRENCE KILHAM Bethesda, Md. (Publication 4259) CITY OF WASHINGTON PUBLISHED BY THE SMITHSONIAN INSTITUTION NOVEMBER 8, 1956 THE LORD BALTIMORE PRESS, INC. BALTIMORE, MD., U. S. A. PREFACE I went to Uganda at the invitation of the East African High Com- mission to carry on virus research as a visiting scientist at the Virus Research Institute, Entebbe, where I worked from August 1954 until mid-May 1955. My ornithological observations were made as an ama- teur in the early mornings and evenings, and on weekends. It had been my hope to study some particular field problem in addition to making a general acquaintance with African bird life. The nature of the prob- lem was determined soon after my arrival. In my bird notes, black- and-white casqued hornbills [Bycanistes suhcylindricits (Sclater)] soon took up more pages than any other species. They came to our garden frequently. In addition, a pair of them roosted and carried on courtship activities in a tree above our house. When I discovered a concentration of hornbill nests in the Mpanga Research Forest, it was apparent that I had an unusual opportunity to study the natural history of casqued hornbills. Present studies did not begin until many females were already walled in. A few pairs of late-nesting hornbills, however, enabled me to witness the beginning stages of nesting ac- tivity. Observations on 16 nesting pairs gave, in the aggregate, a rounded picture of breeding and other habits of these birds. -

P0455-P0459.Pdf
The Condor 101:455-459 0 The Cooper Ornithological Society 1999 BOOK REVIEWS Social Influences on Vocal Development.- Why read the rest of the book if the main themes Charles T Snowdon and Mat-tine Hausberger, eds. are already discussed in the introduction? I believe it 1997. Cambridge University Press, Cambridge, U.K. is worth it, not only to check the statements of the ix + 351 pp. ISBN 0521 49526 1. $95.00 (cloth). editors. All the articles are fine reviews of a given At least in certain aspects, this book is a referee’s field, written by experts. Doug Nelson provides a very dream: not only do the editors provide short summaries good account of the state of the art in song learning in their introduction of the 16 articles making up this theory, and as I expressed above, I fully agree with his volume, they also inform the reader about the main claim that the two stage nature of song learning is still general conclusions that can be drawn from those ar- not fully understood or accepted by other researchers ticles. According to Snowdon and Hausberger, there of the field. are five central themes that arc treated in almost all Luis Baptista and Sandra Gaunt provide a very com- contributions. The first and main theme is the claim petent review on social interaction and vocal devel- that vocal learning has more aspects than learning to opment in birds. As ever, there is a wealth of findings produce sounds. It is also necessary to learn how to from the lab and from the field included in their re- use and comprehend vocalizations. -

Wetlands, Biodiversity and the Ramsar Convention
Wetlands, Biodiversity and the Ramsar Convention Wetlands, Biodiversity and the Ramsar Convention: the role of the Convention on Wetlands in the Conservation and Wise Use of Biodiversity edited by A. J. Hails Ramsar Convention Bureau Ministry of Environment and Forest, India 1996 [1997] Published by the Ramsar Convention Bureau, Gland, Switzerland, with the support of: • the General Directorate of Natural Resources and Environment, Ministry of the Walloon Region, Belgium • the Royal Danish Ministry of Foreign Affairs, Denmark • the National Forest and Nature Agency, Ministry of the Environment and Energy, Denmark • the Ministry of Environment and Forests, India • the Swedish Environmental Protection Agency, Sweden Copyright © Ramsar Convention Bureau, 1997. Reproduction of this publication for educational and other non-commercial purposes is authorised without prior perinission from the copyright holder, providing that full acknowledgement is given. Reproduction for resale or other commercial purposes is prohibited without the prior written permission of the copyright holder. The views of the authors expressed in this work do not necessarily reflect those of the Ramsar Convention Bureau or of the Ministry of the Environment of India. Note: the designation of geographical entities in this book, and the presentation of material, do not imply the expression of any opinion whatsoever on the part of the Ranasar Convention Bureau concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. Citation: Halls, A.J. (ed.), 1997. Wetlands, Biodiversity and the Ramsar Convention: The Role of the Convention on Wetlands in the Conservation and Wise Use of Biodiversity. -

Observations on Rufous-Necked Aceros Nipalensis and Austen's
Observations on Rufous-necked Aceros nipalensis and Austen’s Brown Anorrhinus austeni Hornbills in Arunachal Pradesh: natural history, conservation status, and threats Aparajita Datta Datta, A. 2009. Observations on Rufous-necked Aceros nipalensis and Austen’s Brown Anorrhinus austeni Hornbills in Arunachal Pradesh: natural history, conservation status, and threats. Indian Birds 5 (4): 108–117. Aparajita Datta, Nature Conservation Foundation, 3076/5, 4th Cross, Gokulam Park, Mysore 570002, Karnataka, India. Email: [email protected]. In 1997–1998, Ravi Sankaran had spent three months studying the most interesting, and intriguing, hornbill species found in India, with the smallest global range—the Narcondam Hornbill Aceros narcondami—restricted to a 6 km2 island of the Andaman Islands archipelago. While others before him had spent time on the island and made observations, his were the first systematic and meticulously collected data of a study carried out throughout the breeding season, on a large number of nests. Unfortunately, he never wrote up the work as a publication, but he put his research to good use for conservation action and managed to get the goats that were affecting the regeneration of many hornbill food plants, removed from the island. My paper, in this memorial issue, is about my limited observations on two of the lesser-known, and threatened hornbills of north-eastern India. Abstract Among the five species of hornbills that occur in north-eastern India, the least studied are the endangered Rufous-necked Hornbill Aceros nipalensis, and the Brown Hornbill Anorrhinus austeni1, which has a restricted distribution in India. Based on field surveys conducted in Namdapha National Park, and several forest divisions in eastern Arunachal Pradesh, during 1996–1999 and 2002–2004, I present information on their distribution and relative abundance. -
![Use of Field Recorded Sounds in the Assessment of Forest Birds in Palawan, Philippines ______Required in the Field [19]](https://docslib.b-cdn.net/cover/0725/use-of-field-recorded-sounds-in-the-assessment-of-forest-birds-in-palawan-philippines-required-in-the-field-19-570725.webp)
Use of Field Recorded Sounds in the Assessment of Forest Birds in Palawan, Philippines ______Required in the Field [19]
Asia Pacific Journal of Multidisciplinary Research, Vol. 7, No. 2, May, 2019 _____________________________________________________________________________________________________________________ Asia Pacific Journal of Use of Field Recorded Sounds in the Multidisciplinary Research Assessment of Forest Birds in Palawan, Vol. 7 No.2, 24-31 May 2019 Philippines P-ISSN 2350-7756 E-ISSN 2350-8442 Alejandro A. Bernardo Jr. www.apjmr.com College of Arts and Sciences, Western Philippines University, CHED Recognized Journal Aborlan, Palawan, Philippines ASEAN Citation Index [email protected] Date Received: August 3, 2018; Date Revised: February 8, 2019 Abstract -The uses of bioacoustics in biological applications are getting popular in research communities. Among such application is the use of sound recordings in avifaunal researches. This research explored the possibility of using the sound recording in the assessment of forest birds in Palawan by comparing it in widely used Point Count Method (PCM). To compare the two methods, a simultaneous point count and sound recording surveys from February to November 2017 in the forested slopes of Victoria-Anipahan Mountain in Aborlan, Palawan were conducted. The Sound Recording Method (SRM) listed slightly lower species richness than the PCM, but the difference in the mean number of species was not significant (F1,49=1.05, p > 0.05). The SRM was found to be biased towards noisy and loud calling bird species but it failed to detect the silent and rarely calling species. SRM was also equally sensitive as compared to PCM in detecting endemic and high conservation priority species. Because of these, it was recognized that SRM could be used as one of the alternative methods in forest bird assessment particularly if the concern is avifaunal species richness. -
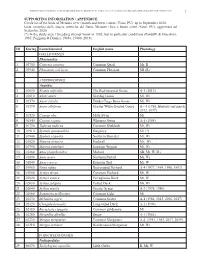
1 ID Euring Latin Binomial English Name Phenology Galliformes
BIRDS OF METAURO RIVER: A GREAT ORNITHOLOGICAL DIVERSITY IN A SMALL ITALIAN URBANIZING BIOTOPE, REQUIRING GREATER PROTECTION 1 SUPPORTING INFORMATION / APPENDICE Check list of the birds of Metauro river (mouth and lower course / Fano, PU), up to September 2020. Lista completa delle specie ornitiche del fiume Metauro (foce e basso corso /Fano, PU), aggiornata ad Settembre 2020. (*) In the study area 1 breeding attempt know in 1985, but in particolar conditions (Pandolfi & Giacchini, 1985; Poggiani & Dionisi, 1988a, 1988b, 2019). ID Euring Latin binomial English name Phenology GALLIFORMES Phasianidae 1 03700 Coturnix coturnix Common Quail Mr, B 2 03940 Phasianus colchicus Common Pheasant SB (R) ANSERIFORMES Anatidae 3 01690 Branta ruficollis The Red-breasted Goose A-1 (2012) 4 01610 Anser anser Greylag Goose Mi, Wi 5 01570 Anser fabalis Tundra/Taiga Bean Goose Mi, Wi 6 01590 Anser albifrons Greater White-fronted Goose A – 4 (1986, february and march 2012, 2017) 7 01520 Cygnus olor Mute Swan Mi 8 01540 Cygnus cygnus Whooper Swan A-1 (1984) 9 01730 Tadorna tadorna Common Shelduck Mr, Wi 10 01910 Spatula querquedula Garganey Mr (*) 11 01940 Spatula clypeata Northern Shoveler Mr, Wi 12 01820 Mareca strepera Gadwall Mr, Wi 13 01790 Mareca penelope Eurasian Wigeon Mr, Wi 14 01860 Anas platyrhynchos Mallard SB, Mr, W (R) 15 01890 Anas acuta Northern Pintail Mi, Wi 16 01840 Anas crecca Eurasian Teal Mr, W 17 01960 Netta rufina Red-crested Pochard A-4 (1977, 1994, 1996, 1997) 18 01980 Aythya ferina Common Pochard Mr, W 19 02020 Aythya nyroca Ferruginous -

Volume 2. Animals
AC20 Doc. 8.5 Annex (English only/Seulement en anglais/Únicamente en inglés) REVIEW OF SIGNIFICANT TRADE ANALYSIS OF TRADE TRENDS WITH NOTES ON THE CONSERVATION STATUS OF SELECTED SPECIES Volume 2. Animals Prepared for the CITES Animals Committee, CITES Secretariat by the United Nations Environment Programme World Conservation Monitoring Centre JANUARY 2004 AC20 Doc. 8.5 – p. 3 Prepared and produced by: UNEP World Conservation Monitoring Centre, Cambridge, UK UNEP WORLD CONSERVATION MONITORING CENTRE (UNEP-WCMC) www.unep-wcmc.org The UNEP World Conservation Monitoring Centre is the biodiversity assessment and policy implementation arm of the United Nations Environment Programme, the world’s foremost intergovernmental environmental organisation. UNEP-WCMC aims to help decision-makers recognise the value of biodiversity to people everywhere, and to apply this knowledge to all that they do. The Centre’s challenge is to transform complex data into policy-relevant information, to build tools and systems for analysis and integration, and to support the needs of nations and the international community as they engage in joint programmes of action. UNEP-WCMC provides objective, scientifically rigorous products and services that include ecosystem assessments, support for implementation of environmental agreements, regional and global biodiversity information, research on threats and impacts, and development of future scenarios for the living world. Prepared for: The CITES Secretariat, Geneva A contribution to UNEP - The United Nations Environment Programme Printed by: UNEP World Conservation Monitoring Centre 219 Huntingdon Road, Cambridge CB3 0DL, UK © Copyright: UNEP World Conservation Monitoring Centre/CITES Secretariat The contents of this report do not necessarily reflect the views or policies of UNEP or contributory organisations. -

Preliminary Report on Wildlife Inventories and Assessment in SFM Project Areas
Preliminary Report on Wildlife Inventories and Assessment in SFM Project Areas Timimbang – Botitian Forest Reserve Prepared by: Rayner Bili Sabah Forestry Department. Survey Period 7th May – 16th May 2014 Date of Report: 18th June 2014 Table of Contents Acknowledgment Abstract List of abbreviations 1.0 INTRODUCTION 1.1 Study Area 1.2 Objectives 2.0 METHODOLOGY 2.1 Recce Walked 2.2 Night Spotting 2.3 Morning Drive 2.4 Camera Trapping 2.5 Interviews 2.6 Opportunistic Sighting 3.0 RESULTS 3.1 Mammals 3.2 Birds 4.0 DISCUSSION 5.0 RECOMMENDATION References Annex I : List of participant and time table Annex II : Datasheet of night spotting Annex III : Datasheet of morning drive Annex IV : Datasheet recce walks Annex V : Opportunistic wildlife sighting sheet Annex VI : Camera trapping datasheet Annex VII : Description of IUCN red list Annex VIII : Photos Acknowledgement By this opportunity, I would like to deeply indebted to Beluran District Forest Officer (DFO) and Assistant District Forest Officers (ADFOs), Forest Rangers, Forester and all forest staff’s of SFM Timimbang-Botitian (Ali Shah Bidin, Mensih Saidin, Jamation Jamion, Jumiting Sauyang and Rozaimee Ahmad) for their help and support during the rapid wildlife survey and assessment in SFM Timimbang-Botitian project area. My sincere thank goes to Mr. Awang Azrul (ADFO) for organizing our accommodation and providing permission to carry out the wildlife survey and for his continuous support for the smooth execution of the programs due the survey requires night movement inside the SFM Timimbang-Botitian forest reserves. Deepest thanks to Mr. Zainal Kula, Mr. Sarinus Aniong and Mr. -

Project Update: July 2010 Sixty Percent of Asia's Forests Are Under High Or Moderate Threat
Project Update: July 2010 Sixty percent of Asia’s forests are under high or moderate threat (Bryant et al., 1997). The main threats for forest loss and degradation are logging, habitat conversion, wildfires, fuel wood collection, overgrazing and plantations. The fundamental triggers behind these activities are probably, increasing human populations, roads penetrating deeper and deeper into the forests and weak economies of respective countries. However, the relative impacts of these factors are still unclear and less apparent. Amongst the causes listed above, logging alone affects a large area of forested tracts in Asia. Logging directly results in change in the structure and composition of forests, in modification of microhabitats and it affects wildlife populations differentially. Responses of different taxa and species within taxa vary to differential pressures of logging. Another serious threat to wildlife populations is hunting. Hunting is widespread across various areas of South and South-east Asia (Bennett et al., 1997; Kinnaird and O'Brien, 2007; Milner-Gulland et al., 2003; Robinson and Bennett, 2000). Rural tribal communities hunt for various reasons from meeting dietary needs and protein requirements, to hunting specific species for valuable, culturally significant, or medicinally important body parts. Asian hornbills are a group of large birds, which are restricted to the tropical forests of South and South-east Asia. There are 31 species of hornbills. Hornbills face severe threats from both logging and hunting. Unlike many other hole-nesting birds, hornbills are secondary cavity-nesters, which mean they cannot make their own cavity. They require large cavities on tall emergent large trees especially because the female incarcerates herself in the nest during the breeding season. -

Seed Dispersal by Ceratogymna Hornbills in the Dja Reserve, Cameroon
Journal of Tropical Ecology (1998) 14:351–371. With 2 figures Copyright 1998 Cambridge University Press Seed dispersal by Ceratogymna hornbills in the Dja Reserve, Cameroon KENNETH D. WHITNEY*†1, MARK K. FOGIEL*, AARON M. LAMPERTI*, KIMBERLY M. HOLBROOK*, DONALD J. STAUFFER*, BRITTA DENISE HARDESTY*, V. THOMAS PARKER* and THOMAS B. SMITH*† *Department of Biology, San Francisco State University, 1600 Holloway Avenue, San Francisco, California 94132 U.S.A. †Center for Population Biology, University of California, Davis, California 95616-8755 U.S.A. (Accepted 13 January 1998) ABSTRACT. Seed dispersal is a process critical to the maintenance of tropical forests, yet little is known about the interactions of most dispersers with their communities. In the Dja Reserve, Cameroon, seed dispersal by the hornbills Cerato- gymna atrata, C. cylindricus and C. fistulator (Aves: Bucerotidae) was evaluated with respect to the taxonomic breadth of plants dispersed, location of seed deposition and effects on seed germination. Collectively, the three hornbill species consumed fruits from 59 tree and liana species, and likely provided dispersal for 56 of them. Hornbill-dispersed tree species composed 22% of the known tree flora of the site. Hornbill visit lengths, visit frequencies, and seed passage times indicated that few seeds were deposited beneath parent trees; in five hornbill/tree species pairings studied, 69–100% of the seeds ingested were deposited away from the parent trees. Germination trials showed that hornbill gut passage is gentle on seeds. Of 24 tree species tested, 23 germinated after passage by hornbills; of 17 planted with con- trols taken directly from trees, only four species showed evidence of inhibition of germination rate, while seven experienced unchanged germination rates and six experienced enhanced germination rates. -

Visayan Hornbill
Visayan hornbill The Visayan hornbill (Penelopides panini) is a hornbill found in rainforests on the islands of Panay, Negros, Masbate, and Visayan hornbill Guimaras, and formerly Ticao, in the Philippines. It formerly included all other Philippine tarictic hornbills as subspecies, in which case the common name of the 'combined species' was shortened to tarictic hornbill. Contents Taxonomy Description Diet and behavior Pair at Avifauna in Alphen aan den Conservation Rijn, Netherlands. Captivity Conservation status References External links Endangered (IUCN 3.1)[1] Taxonomy Scientific classification The Visayan hornbill was described by the French polymath Kingdom: Animalia Georges-Louis Leclerc, Comte de Buffon in 1780 in his Histoire Phylum: Chordata Naturelle des Oiseaux.[2] The bird was also illustrated in a hand- coloured plate engraved by François-Nicolas Martinet in the Class: Aves Planches Enluminées D'Histoire Naturelle which was produced under the supervision of Edme-Louis Daubenton to accompany Order: Bucerotiformes Buffon's text.[3] Neither the plate caption nor Buffon's description Family: Bucerotidae included a scientific name but in 1783 the Dutch naturalist Pieter Boddaert coined the binomial name Buceros panini in his catalogue Genus: Penelopides of the Planches Enluminées.[4] The type locality is the island of Species: P. panini Panay in the Philippines.[5] The Visayan hornbill is now placed in Binomial name the genus Penelopides that was introduced in 1849 by the German naturalist Ludwig Reichenbach in a plate of the hornbills.[6][7] The Penelopides panini origin of the generic name is uncertain but it may be a combination (Boddaert, 1783) of the Latin pene meaning "almost" or "nearly", the Ancient Greek lophos meaning "crest" and -oideēs "resembling".