Approximation Algorithms Under the Worst-Case Analysis and the Smoothed Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Smoothed Possibility of Social Choice
The Smoothed Possibility of Social Choice Lirong Xia, RPI, [email protected] Abstract We develop a framework that leverages the smoothed complexity analysis by Spielman and Teng [60] to circumvent paradoxes and impossibility theorems in social choice, motivated by modern applications of social choice powered by AI and ML. For Condrocet’s paradox, we prove that the smoothed likelihood of the paradox either vanishes at an exponential rate as the number of agents in- creases, or does not vanish at all. For the ANR impossibility on the non-existence of voting rules that simultaneously satisfy anonymity, neutrality, and resolvability, we characterize the rate for the impossibility to vanish, to be either polynomially fast or exponentially fast. We also propose a novel easy-to-compute tie-breaking mechanism that optimally preserves anonymity and neutrality for even number of alternatives in natural settings. Our results illustrate the smoothed possibility of social choice—even though the paradox and the impossibility theorem hold in the worst case, they may not be a big concern in practice. 1 Introduction Dealing with paradoxes and impossibility theorems is a major challenge in social choice theory, because “the force and widespread presence of impossibility results generated a consolidated sense of pessimism, and this became a dominant theme in welfare economics and social choice theory in general”, as the eminent economist Amartya Sen commented in his Nobel prize lecture [57]. Many paradoxes and impossibility theorems in social choice are based on worst-case analysis. Take perhaps the earliest one, namely Condorcet’s (voting) paradox [15], for example. Condorcet’s para- dox states that, when there are at least three alternatives, it is impossible for pairwise majority aggregation to be transitive. -

Smoothed Complexity Theory And, Thus, to Embed Smoothed Analysis Into Compu- Tational Complexity
Smoothed Complexity Theory∗ Markus Bl¨aser1 Bodo Manthey2 1Saarland University, Department of Computer Science, [email protected] 2University of Twente, Department of Applied Mathematics, [email protected] Smoothed analysis is a new way of analyzing algorithms introduced by Spielman and Teng (J. ACM, 2004). Classical methods like worst-case or average-case anal- ysis have accompanying complexity classes, like P and Avg-P, respectively. While worst-case or average-case analysis give us a means to talk about the running time of a particular algorithm, complexity classes allows us to talk about the inherent difficulty of problems. Smoothed analysis is a hybrid of worst-case and average-case analysis and com- pensates some of their drawbacks. Despite its success for the analysis of single algorithms and problems, there is no embedding of smoothed analysis into com- putational complexity theory, which is necessary to classify problems according to their intrinsic difficulty. We propose a framework for smoothed complexity theory, define the relevant classes, and prove some first hardness results (of bounded halting and tiling) and tractability results (binary optimization problems, graph coloring, satisfiability). Furthermore, we discuss extensions and shortcomings of our model and relate it to semi-random models. 1 Introduction The goal of computational complexity theory is to classify computational problems according to their intrinsic difficulty. While the analysis of algorithms is concerned with analyzing, say, the running time of a particular algorithm, complexity theory rather analyses the amount of resources that all algorithms need at least to solve a given problem. Classical complexity classes, like P, reflect worst-case analysis of algorithms. -

Smoothed Analysis of the 2-Opt Heuristic for the TSP: Polynomial Bounds for Gaussian Noise∗
Smoothed Analysis of the 2-Opt Heuristic for the TSP: Polynomial Bounds for Gaussian Noise∗ Bodo Manthey Rianne Veenstra University of Twente, Enschede, Netherlands, Department of Applied Mathematics [email protected], [email protected] The 2-opt heuristic is a very simple local search heuristic for the traveling sales- man problem. While it usually converges quickly in practice, its running-time can be exponential in the worst case. Englert, R¨oglin,and V¨ocking (Algorithmica, 2014) provided a smoothed analysis in the so-called one-step model in order to explain the performance of 2-opt on d- dimensional Euclidean instances. However, translating their results to the classical model of smoothed analysis, where points are perturbed by Gaussian distributions with standard deviation σ, yields only bounds that are polynomial in n and 1/σd. We prove bounds that are polynomial in n and 1/σ for the smoothed running- time with Gaussian perturbations. In addition, our analysis for Euclidean distances is much simpler than the existing smoothed analysis. 1 2-Opt and Smoothed Analysis The traveling salesman problem (TSP) is one of the classical combinatorial optimization prob- lems. Euclidean TSP is the following variant: given points X ⊆ [0; 1]d, find the shortest Hamiltonian cycle that visits all points in X (also called a tour). Even this restricted variant is NP-hard for d ≥ 2 [21]. We consider Euclidean TSP with Manhattan and Euclidean dis- tances as well as squared Euclidean distances to measure the distances between points. For the former two, there exist polynomial-time approximation schemes (PTAS) [2, 20]. -

Smoothed Analysis of the TSP Algorithms
Smoothed Analysis of the TSP Algorithms Jens Ziegler Geboren am 11. Februar 1989 in Siegburg September 22, 2012 Bachelorarbeit Mathematik Betreuer: Prof. Dr. Nitin Saxena Mathematisches Institut Mathematisch-Naturwissenschaftliche Fakultat¨ der Rheinischen Friedrich-Wilhelms-Universitat¨ Bonn Contents 1 Introduction 2 2 Preliminaries 6 3 2-Opt 8 3.1 Expected Number of 2-Changes on the L1 Metric . 8 3.1.1 Pairs of Linked 2-Changes . 10 3.1.2 Analysing the Pairs of Linked 2-Changes . 12 3.1.3 Proofs of Theorem 1 a) and Theorem 2 a) . 16 3.2 Expected Number of 2-Changes on the L2 Metric . 17 3.2.1 Analysis of a Single 2-Change . 18 3.2.2 Simplified Random Experiments . 29 3.2.3 Analysis of Pairs of Linked 2-Changes . 31 3.2.4 ProofsofTheorem1b)and2b) . 34 3.3 Expected Number of 2-Changes on General Graphs . 36 3.3.1 Definition of Witness Sequences . 36 3.3.2 Improvement to the Tour Made by Witness Sequences 37 3.3.3 Identifying Witness Sequences . 39 3.3.4 ProofofTheorem1c)and2c) . 43 3.4 ProofofTheorem3........................ 44 3.5 SmoothedAnalysisof2-Opt. 46 4 Karp’s Partitioning Scheme 48 4.1 Preliminaries ........................... 48 4.2 NecessaryLemmasandTheorems. 49 4.3 Smoothed Analysis of Karp’s Partitioning Scheme . 52 5 Conclusions and Open Questions 54 1 1 Introduction Smoothed Analysis was first introduced by Daniel A. Spielman and Shang- Hua Teng in 2001. Up until that time, there were only two standard ways of measuring an algorithms complexity: worst-case and average-case-analysis. -

Smoothed Analysis of Local Search Algorithms
Smoothed Analysis of Local Search Algorithms Bodo Manthey University of Twente, Department of Applied Mathematics Enschede, The Netherlands, [email protected] Abstract. Smoothed analysis is a method for analyzing the perfor- mance of algorithms for which classical worst-case analysis fails to ex- plain the performance observed in practice. Smoothed analysis has been applied to explain the performance of a variety of algorithms in the last years. One particular class of algorithms where smoothed analysis has been used successfully are local search algorithms. We give a survey of smoothed analysis, in particular applied to local search algorithms. 1 Smoothed Analysis 1.1 Motivation The goal of the analysis of algorithms is to provide measures for the performance of algorithms. In this way, it helps to compare algorithms and to understand their behavior. The most commonly used method for the performance of algorithms is worst-case analysis. If an algorithm has a good worst-case performance, then this is a very strong statement and, up to constants and lower order terms, the algorithm should also perform well in practice. However, there are many algorithms that work surprisingly well in practice although they have a very poor worst-case performance. The reason for this is that the worst-case performance can be dominated by a few pathological instances that hardly or never occur in practice. A frequently used alternative to worst-case analysis is average-case analysis. In average-case analysis, the expected performance is measured with respect to some fixed probability distribution. Many algorithms with poor worst-case but good practical performance show a good average-case performance. -

Smoothed Analysis of Algorithms, We Measure the Expected Performance of Algorithms Under Slight Random Perturbations of Worst-Case Inputs
ICM 2002 Vol. I 597–606 · · Smoothed Analysis of Algorithms Daniel A. Spielman∗ Shang-Hua Teng† Abstract Spielman and Teng [STOC ’01] introduced the smoothed analysis of al- gorithms to provide a framework in which one could explain the success in practice of algorithms and heuristics that could not be understood through the traditional worst-case and average-case analyses. In this talk, we survey some of the smoothed analyses that have been performed. 2000 Mathematics Subject Classification: 65Y20, 68Q17, 68Q25, 90C05. Keywords and Phrases: Smoothed analysis, Simplex method, Condition number, Interior point method, Perceptron algorithm. 1. Introduction The most common theoretical approach to understanding the behavior of al- gorithms is worst-case analysis. In worst-case analysis, one proves a bound on the worst possible performance an algorithm can have. A triumph of the Algo- rithms community has been the proof that many algorithms have good worst-case performance—a strong guarantee that is desirable in many applications. However, there are many algorithms that work exceedingly well in practice, but which are known to perform poorly in the worst-case or lack good worst-case analyses. In an attempt to rectify this discrepancy between theoretical analysis and observed performance, researchers introduced the average-case analysis of algorithms. In average-case analysis, one bounds the expected performance of an algorithm on arXiv:math/0212413v1 [math.OC] 1 Dec 2002 random inputs. While a proof of good average-case performance provides evidence that an algorithm may perform well in practice, it can rarely be understood to ex- plain the good behavior of an algorithm in practice. -

Exploring Parameter Spaces in Coping with Computational Intractability
Exploring Parameter Spaces in Coping with Computational Intractability Vorgelegt von Diplom-Informatiker Sepp Hartung geboren in Jena, Thüringen Von der Fakultät IV – Elektrotechnik und Informatik der Technischen Universität Berlin zur Erlangung des akademischen Grades Doktor der Naturwissenschaften – Dr. rer. nat. – genehmigte Dissertation Promotionsausschuss: Vorsitzender: Prof. Dr. Stephan Kreutzer Gutachter: Prof. Dr. Rolf Niedermeier Gutachter: Prof. Dr. Pinar Heggernes Gutachter: Prof. Dr. Dieter Kratsch Tag der wissenschaftlichen Aussprache: 06. Dezember 2013 Berlin 2014 D83 Zusammenfassung In dieser Arbeit werden verschiedene Ansätze zur Identifikation von effizient lösbaren Spezialfällen für schwere Berechnungsprobleme untersucht. Dabei liegt der Schwerpunkt der Betrachtungen auf der Ermittlung des Einflusses sogenannter Problemparameter auf die Berechnungsschwere. Für Betrach- tungen dieser Art bietet die parametrisierte Komplexitätstheorie unsere theoretische Basis. In der parametrisierten Komplexitätstheorie wird versucht sogenannte Pro- blemparameter in schweren Berechnungsproblemen (meistens, gehören diese zu den NP-schweren Problemen) zu identifizieren und diese zur Entwicklung von parametrisierten Algorithmen zu nutzen. Parametrisierte Algorithmen sind dadurch gekennzeichnet, dass der exponentielle Laufzeitanteil durch eine Funktion abhängig allein vom Problemparameter beschränkt werden kann. Die Entwicklung von parametrisierten Algorithmen ist stark motiviert durch sogenannte heuristische Algorithmen. Entgegen der theoretisch -

Beyond Worst-Case Analysis Lecture #1: Three Motivating Examples∗
CS264: Beyond Worst-Case Analysis Lecture #1: Three Motivating Examples∗ Tim Roughgardeny September 22, 2014 1 Preamble We begin in Sections 2{4 with three examples that clearly justify the need for a course like this, and for alternatives to the traditional worst-case analysis of algorithms. Our discussion is brief and informal, to provide motivation and context. Later lectures treat these examples, and many others, in depth. 2 Caching: LRU vs. FIFO Our first example demonstrates how the worst-case analysis of algorithms can fail to differ- entiate between empirically \better" and \worse" solutions. The setup should be familiar from your system courses, if not your algorithms courses. There is a small fast memory (the \cache") and a big slow memory. For example, the former could be an on-chip cache and the latter main memory; or the former could be main memory and the latter a disk. There is a program that periodically issues read and write requests to data stored on \pages" in the big slow memory. In the happy event that the requested page is also in the cache, then it can be accessed directly. If not | on a \page fault" a.k.a. \cache miss" | the requested page needs to be brought into the cache, and this requires evicting an incumbent page. The key algorithmic question is then: which page should be evicted? For example, consider a cache that has room for four pages (Figure 1). Suppose the first four page requests are for a, b, c, and d; these are brought into the (initially empty) cache in succession. -

Professor Shang-Hua Teng
Professor Shang-Hua Teng Department of Computer Science, University of Southern California 1039 25 Street 3710 McClintock, RTH 505, Los Angeles, CA 90089 Santa Monica, CA 90403 (213) 740-3667, [email protected] (617) 440-4281 EMPLOYMENT: University Professor USC (2017 – ) Seeley G. Mudd Professor (Computer Science and Mathematics) USC (2012 – ) Chair and Seeley G. Mudd Professor (Computer Science) USC (2009 – 2012) Professor (Computer Science) Boston University (2002 – 2009) Research Affiliate Professor (Mathematics) MIT (1999 – Present) Visiting Research Scientist Microsoft Research Asia and New England (2004 – Present) Senior Research Scientist Akamai (1997 – 2009) Visiting Professor (Computer Science) Tsinghua University (2004 – 2008) Professor (Computer Science)[Associate Professor before 1999] UIUC (2000 – 2002) Research Scientist IBM Almaden Research Center (1997 – 1999) Assistant Professor (Computer Science) Univ. of Minnesota (1994 – 1997) Research Scientist Intel Corporation (Summer 1994, 1995) Instructor (Mathematics) MIT (1992 – 1994) Computer Scientist NASA Ames Research Center (Summer 1993) Research Scientist Xerox Palo Alto Research Center (1991 – 1992) EDUCATION: Ph.D. (Computer Science) Carnegie Mellon University (1991) Thesis: “A Unified Geometric Approach to Graph Partitioning.” Advisor: Gary Lee Miller M.S. (Computer Science) University of Southern California (1988) Thesis: “Security, Verifiablity, and Universality in Distributed Computing”. Advised by Leonard Adleman and Ming-Deh Huang B.S. (Computer Science) & B.A. (Electrical -
1 Introduction
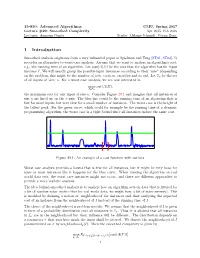
15-850: Advanced Algorithms CMU, Spring 2017 Lecture #29: Smoothed Complexity Apr 2015, Feb 2016 Lecturer: Anupam Gupta Scribe: Melanie Schmidt, Goran Zuzic 1 Introduction Smoothed analysis originates from a very influential paper of Spielman and Teng [ST01, ST04]. It provides an alternative to worst-case analysis. Assume that we want to analyze an algorithm's cost, e.g., the running time of an algorithm. Let cost(A(I)) be the cost that the algorithm has for input instance I. We will usually group the possible input instances according to their \size" (depending on the problem, this might be the number of jobs, vertices, variables and so on). Let In be the set of all inputs of `size' n. For a worst case analysis, we are now interested in max cost(A(I)); I2In the maximum cost for any input of size n. Consider Figure 29.1 and imagine that all instances of size n are lined up on the x-axis. The blue line could be the running time of an algorithm that is fast for most inputs but very slow for a small number of instances. The worst case is the height of the tallest peak. For the green curve, which could for example be the running time of a dynamic programming algorithm, the worst case is a tight bound since all instances induce the same cost. Figure 29.1: An example of a cost function with outliers. Worst case analysis provides a bound that is true for all instances, but it might be very loose for some or most instances like it happens for the blue curve. -

Patterns, Predictions, and Actions
PATTERNS, PREDICTIONS, AND ACTIONS A story about machine learning Moritz Hardt and Benjamin Recht arXiv:2102.05242v1 [cs.LG] 10 Feb 2021 Working draft Created: Tue 09 Feb 2021 07:25:15 PM PST Latest version available at https://mlstory.org. For Isaac, Leonora, and Quentin Contents Introduction 9 Ambitions of the 20th century 10 Pattern classification 12 Prediction and action 16 About this book 18 Chapter notes 20 Acknowledgments 21 Decision making 23 Bayesian binary hypothesis testing 25 Canonical cases of likelihood ratio tests 28 Types of errors and successes 29 The Neyman-Pearson Lemma 30 Properties of ROC curves 32 Looking ahead: what if we don’t know the models? 33 Decisions that discriminate 34 Chapter notes 38 Supervised learning 41 Sample versus population 42 Supervised learning 43 A first learning algorithm: The perceptron 45 Chapter notes 53 6 Representations and features 55 Measurement 56 Quantization 57 Template matching 59 Summarization and histograms 60 Nonlinear predictors 61 Chapter notes 72 Optimization 75 Optimization basics 76 Gradient descent 77 Applications to empirical risk minimization 80 Insights on convergence of gradient methods from quadratic functions 82 Stochastic gradient descent 84 Analysis of the stochastic gradient method 90 Implicit convexity 92 Regularization 96 Squared loss methods and other optimization tools 100 Chapter notes 101 Generalization 103 Generalization gap 103 Overparameterization: empirical phenomena 104 Theories of generalization 108 Algorithmic stability 110 Model complexity and uniform -

Beyond Worst-Case Analysis
Beyond Worst-Case Analysis Tim Roughgarden∗ June 27, 2018 1 Introduction Comparing different algorithms is hard. For almost any pair of algorithms and measure of algorithm performance|like running time or solution quality|each algorithm will perform better than the other on some inputs.1 For example, the insertion sort algorithm is faster than merge sort on already-sorted arrays but slower on many other inputs. When two algorithms have incomparable performance, how can we deem one of them \better than" the other? Worst-case analysis is a specific modeling choice in the analysis of algorithms, where the overall performance of an algorithm is summarized by its worst performance on any input of a given size. The \better" algorithm is then the one with superior worst-case performance. Merge sort, with its worst-case asymptotic running time of Θ(n log n) for arrays of length n, is better in this sense than insertion sort, which has a worst-case running time of Θ(n2). While crude, worst-case analysis can be tremendously useful, and it is the dominant paradigm for algorithm analysis in theoretical computer science. A good worst-case guarantee is the best- case scenario for an algorithm, certifying its general-purpose utility and absolving its users from understanding which inputs are relevant to their applications. Remarkably, for many fundamental computational problems, there are algorithms with excellent worst-case performance guarantees. The lion's share of an undergraduate algorithms course comprises algorithms that run in linear or near-linear time in the worst case. For many problems a bit beyond the scope of an undergraduate course, however, the downside of worst-case analysis rears its ugly head.