Download/ (Accessed on 3 May 2020)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amazing Facts the King Vulture Is the Only Surviving Member of the Genus Sarcoramphus
King Vulture Sarcoramphus papa Bird Scientific Name Sarcoramphus papa Other Names None Range South and Central America from Mexico to northern Argentina Habitat Savannas, tropical forests and grasslands Description A large, white vulture with grayish-black wings and tail feathers, and a pronounced skin flap (caruncle) on the upper beak. The head and neck are bald with the skin color varying in Behavior color including yellow, orange, blue, purple, These birds are diurnal, spending their daytime hours soaring high in and red. the sky on thermals searching for food using keen eyesight. They are Average Size considered resident birds that are non-migratory and maintain a set Height: 20 in. home range. King Vultures are often seen alone or in small family groups Wingspan: 5.5 – 6.5 ft. numbering approximately 15 individuals, and will tolerate other animals and Weight: 6 – 10 lbs. vulture species in the same proximity. Even though they are the dominant vulture at the feeding site and usually the first to arrive, they are not Lifespan territorial with the smaller vultures that feed alongside them. In the wild: Estimated at 25 years The head and neck of the King Vulture lack feathers to help prevent In captivity: Up to 35 years infections and to keep the remains of the carcass from damaging the feathers. After eating, Vultures relax in the sun and allow the heat to bake Diet off the bacteria. In the wild: Carrion In captivity: Rodents, fish and prepared meat Reproduction and Breeding Incubation There is limited information on wild courtship and breeding of this species, 53 – 58 days but because they breed well in captivity, there is reliable data from numerous captive settings. -

A Multi-Gene Phylogeny of Aquiline Eagles (Aves: Accipitriformes) Reveals Extensive Paraphyly at the Genus Level
Available online at www.sciencedirect.com MOLECULAR SCIENCE•NCE /W\/Q^DIRI DIRECT® PHYLOGENETICS AND EVOLUTION ELSEVIER Molecular Phylogenetics and Evolution 35 (2005) 147-164 www.elsevier.com/locate/ympev A multi-gene phylogeny of aquiline eagles (Aves: Accipitriformes) reveals extensive paraphyly at the genus level Andreas J. Helbig'^*, Annett Kocum'^, Ingrid Seibold^, Michael J. Braun^ '^ Institute of Zoology, University of Greifswald, Vogelwarte Hiddensee, D-18565 Kloster, Germany Department of Zoology, National Museum of Natural History, Smithsonian Institution, 4210 Silver Hill Rd., Suitland, MD 20746, USA Received 19 March 2004; revised 21 September 2004 Available online 24 December 2004 Abstract The phylogeny of the tribe Aquilini (eagles with fully feathered tarsi) was investigated using 4.2 kb of DNA sequence of one mito- chondrial (cyt b) and three nuclear loci (RAG-1 coding region, LDH intron 3, and adenylate-kinase intron 5). Phylogenetic signal was highly congruent and complementary between mtDNA and nuclear genes. In addition to single-nucleotide variation, shared deletions in nuclear introns supported one basal and two peripheral clades within the Aquilini. Monophyly of the Aquilini relative to other birds of prey was confirmed. However, all polytypic genera within the tribe, Spizaetus, Aquila, Hieraaetus, turned out to be non-monophyletic. Old World Spizaetus and Stephanoaetus together appear to be the sister group of the rest of the Aquilini. Spiza- stur melanoleucus and Oroaetus isidori axe nested among the New World Spizaetus species and should be merged with that genus. The Old World 'Spizaetus' species should be assigned to the genus Nisaetus (Hodgson, 1836). The sister species of the two spotted eagles (Aquila clanga and Aquila pomarina) is the African Long-crested Eagle (Lophaetus occipitalis). -

Application for Non-Resident Raptor Trapping Permit
Application for Non-Resident Raptor Trapping Permit Please complete this form and return it, along with the permit fee and requested attachments, to: Texas Parks and Wildlife Department, Wildlife Diversity Permits, 4200 Smith School Road, Austin, TX 78744. All applicants for a Non-Resident Raptor Trapping Permit must possess a permit from their home state equivalent to a Texas Apprentice, General or Master Falconer permit. A non-resident shall not st th trap more than one raptor per year in this state (July 1 – June 30 ). Applicant Name: Home Phone: Office Phone: Street Address: Cell Phone: Driver's License No.: City: State: Zip: Date of Birth: 1 Social Security E-mail Address: Number: 1. This application is for a Non-Resident Raptor Trapping Permit to allow a permitted non-resident apprentice, general or master falconer to take, from the wild in Texas with a valid non-resident hunting license, one of the following species of raptors within the permit year (July 1st – June 30th): Sharp-shinned hawk (Accipiter striatus), Cooper’s hawk (Accipiter cooperii), Harris’s Hawk (Parabuteo unicinctus), Red-shouldered hawk (Buteo lineatus), Red-tailed hawk (Buteo jamaicensis), Ferruginous hawk (Buteo regalis), American kestrel (Falco sparverius), Merlin (Falco columbarius), Prairie falcon (Falco mexicanus), Gyrfalcon (Falco rusticolus), or Caracara (Caracara cheriway). 2. Please attach a copy of your current state falconry permit. 3. Please enclose the $378.00 application and permit fee. (Make checks or money order payable to Texas Parks and Wildlife Department) The Texas Administrative Code Raptor Proclamation can be found online at: http://texreg.sos.state.tx.us/public/readtac$ext.ViewTAC?tac_view=5&ti=31&pt=2&ch=65&sch=K&rl=Y I have read and understand the state regulations titled “Raptor Proclamation” and I will comply with all state and federal laws pertaining to falconry. -

Chromosome Painting in Three Species of Buteoninae: a Cytogenetic Signature Reinforces the Monophyly of South American Species
Chromosome Painting in Three Species of Buteoninae: A Cytogenetic Signature Reinforces the Monophyly of South American Species Edivaldo Herculano C. de Oliveira1,2,3*, Marcella Mergulha˜o Tagliarini4, Michelly S. dos Santos5, Patricia C. M. O’Brien3, Malcolm A. Ferguson-Smith3 1 Laborato´rio de Cultura de Tecidos e Citogene´tica, SAMAM, Instituto Evandro Chagas, Ananindeua, PA, Brazil, 2 Faculdade de Cieˆncias Exatas e Naturais, ICEN, Universidade Federal do Para´, Bele´m, PA, Brazil, 3 Cambridge Resource Centre for Comparative Genomics, Cambridge, United Kingdom, 4 Programa de Po´s Graduac¸a˜oem Neurocieˆncias e Biologia Celular, ICB, Universidade Federal do Para´, Bele´m, PA, Brazil, 5 PIBIC – Universidade Federal do Para´, Bele´m, PA, Brazil Abstract Buteoninae (Falconiformes, Accipitridae) consist of the widely distributed genus Buteo, and several closely related species in a group called ‘‘sub-buteonine hawks’’, such as Buteogallus, Parabuteo, Asturina, Leucopternis and Busarellus, with unsolved phylogenetic relationships. Diploid number ranges between 2n = 66 and 2n = 68. Only one species, L. albicollis had its karyotype analyzed by molecular cytogenetics. The aim of this study was to present chromosomal analysis of three species of Buteoninae: Rupornis magnirostris, Asturina nitida and Buteogallus meridionallis using fluorescence in situ hybridization (FISH) experiments with telomeric and rDNA probes, as well as whole chromosome probes derived from Gallus gallus and Leucopternis albicollis. The three species analyzed herein showed similar karyotypes, with 2n = 68. Telomeric probes showed some interstitial telomeric sequences, which could be resulted by fusion processes occurred in the chromosomal evolution of the group, including the one found in the tassociation GGA1p/GGA6. -

Trade in Andean Condor Vulture Gryphus Feathers and Body Parts in the City of Cusco and the Sacred Valley, Cusco Region, Peru Robert S
Vulture News 61 September 2011 Trade in Andean Condor Vulture gryphus feathers and body parts in the city of Cusco and the Sacred Valley, Cusco region, Peru Robert S. R. Williams1*, Jose Luis Jara1, Daphne Matsufuiji2 and Anahi Plenge2 1Frankfurt Zoological Society and the Andean Condor Working Group – Peru 2Clorinda Matto de Turner 305, Urb. Magisterio, Cusco, Peru *Corresponding author: [email protected] Summary The sale of Andean Condor feathers and body parts is undertaken openly in the tourist markets of Cusco and the Sacred Valley. This trade is illegal but there is no enforcement of existing legislation. We visited the main tourist markets of the region to ascertain the extent of the trade, reasons motivating it and value. We found condor feathers for sale in 26 establishments. Feathers were sold singly, decorated and incorporated in handicrafts. Prices ranged from 5 soles for a small body feather to 160 soles for a main primary and we found handicrafts for sale at prices of up to 650 soles (featuring 6 feathers). We were offered a whole condor for sale at a market in Cusco for 2,500 soles. Investigations revealed that there are condor hunters working to supply this trade in both the Cordillera Vilcabamba and Cordillera Vilcanota and that the town of Calca is the base of much of the handicraft production. The trade is mainly based on three uses: alternative healing, shamanic ceremonies and souvenirs. It is crucial that the Peruvian authorities honour their commitments under international conventions and act immediately to stop this illegal trade, which is further threatening a species that is already in a precarious situation. -

Bald Eagle Haliaeetus Leucocephalus
Wyoming Species Account Bald Eagle Haliaeetus leucocephalus REGULATORY STATUS USFWS: Delisted; Migratory Bird USFS R2: Sensitive USFS R4: Sensitive Wyoming BLM: Sensitive State of Wyoming: Protected Bird CONSERVATION RANKS USFWS: Bird of Conservation Concern WGFD: NSS3 (Bb), Tier II WYNDD: G5, S4B/S5N Wyoming Contribution: LOW IUCN: Least Concern PIF Continental Concern Score: 9 STATUS AND RANK COMMENTS Bald Eagle (Haliaeetus leucocephalus) is provided international protection under the Federal Migratory Bird Treaty Act of 1918, as amended 1. In 1940, Bald Eagle was provided protection under the Bald and Golden Eagle Protection Act 2. In 1966, the southern subspecies was listed as federally endangered under the Endangered Species Preservation Act; the entire population in the contiguous United States was listed as endangered in 1978 under the 1973 Endangered Species Act (ESA). A significant increase in numbers of nesting pairs, productivity, and distribution allowed Bald Eagle to be reclassified from Endangered to Threatened in 1995 under the ESA 3. Bald Eagle was delisted in 2007, and numbers are considered to be stable to increasing across its range 4. The species has been assigned different state conservation ranks by the Wyoming Natural Diversity Database for the breeding season and nonbreeding season because the abundance of the species is different between seasons. NATURAL HISTORY Taxonomy: Bald Eagle is a member of the family Accipitridae, which includes kites, eagles, harriers, and hawks 5. There are two subspecies of Bald Eagle; H. l. alascanus is found north of 40 degrees latitude across North America, including Wyoming, while H. l. leucocephalus is found south of 40 degrees latitude in the Gulf coast states 6. -

Regional Specialties Western
REGIONAL SPECIALTIES WESTERN OSPREY 21 - 26” length SOUTHERN . FERRUGINOUS . Eagle sized; clean, white body. HAWK Black wrist marks. 20 - 26” length . Glides with kink (M) in long, narrow wings. MISSISSIPPI . Largest buteo; eagle-like. KITE . Pale below with dark leggings. 13 - 15” length . Mostly white tail; 3 color morphs. Long, pointed wings; slim body. Light body; dark wings; narrow, black tail. Not to scale. Buoyant, acrobatic flight. NORTHERN HARRIER 16 - 20” length PRAIRIE FALCON 14 - 18” length . Long, narrow wings and tail; sharp dihedral. Size of Peregrine; much paler plumage. Brown above, streaked brown below – female. Narrow moustache; spotted breast; long tail. Gray above, pale below with black wing tips – male. Dark armpits and partial wing linings. WING PROFILE IMMATURE BALD EAGLE BALD EAGLE GOLDEN EAGLE . Immature birds vary GOLDEN EAGLE greatly in the amount 27 to 35” length of white spotting on body and wings. White showing on wing linings is surely a Bald Eagle. BALD EAGLE . Like large buteo, curvy wings. Head protrudes much less than tail. Slight dihedral to wing profile. NOTE: Some hawks soar and glide with their wings raised above the horizontal, called a dihedral. 27 to 35” length . Head and tail length similar. Long, flat wings. Straight leading edge to wings. 24 to 28” length This guide developed by Paul Carrier is the property of the Hawk Migration Association of North America (HMANA). HMANA is TURKey VUltUre a membership-based, non-profit organization committed to the . Dark wing linings with light flight feathers. conservation of raptors through the scientific study, enjoyment, and . Small head; long tail; sharp dihedral. -

Birds of Prey (Accipitriformes and Falconiformes) of Serra De Itabaiana National Park, Northeastern Brazil
Acta Brasiliensis 4(3): 156-160, 2020 Original Article http://revistas.ufcg.edu.br/ActaBra http://dx.doi.org/10.22571/2526-4338416 Birds of prey (Accipitriformes and Falconiformes) of Serra de Itabaiana National Park, Northeastern Brazil Cleverton da Silvaa* i , Cristiano Schetini de Azevedob i , Juan Ruiz-Esparzac i , Adauto de Souza d i Ribeiro h a Programa de Pós-Graduação em Desenvolvimento e Meio Ambiente, Universidade Federal de Sergipe, Aracajú, São Cristóvão, 49100-100, Sergipe, Brasil. *[email protected] b Programa de Pós-Graduação em Ecologia de Biomas Tropicais, Universidade Federal de Ouro Preto, Ouro Preto, 35400-000, Minas Gerais, Brasil. c Universidade Federal de Sergipe, Nossa Senhora da Glória, 49680-000, Sergipe, Brasil. d Universidade Federal de Sergipe, Aracajú, São Cristóvão, 49100-100, Sergipe, Brasil. Received: June 20, 2020 / Accepted: August 27, 2020/ Published online: September 28, 2020 Abstract Birds of prey are important for maintaining ecosystems, since they can regulate the populations of vertebrates and invertebrates. However, anthropic activities, like habitat fragmentation, have been decreasing the number of birds of prey, affecting the habitat ecological relations and, decreasing biodiversity. Our objective was to evaluate species of birds of prey (Accipitriformes and Falconiformes) in a protected area of the Atlantic forest in northeastern Brazil. The area was sampled for 17 months using fixed points and walking along a pre-existing trail. Birds of prey were classified by their Punctual Abundance Index, threat status and forest dependence. Sixteen birds of prey were recorded, being the most common Rupornis magnirostris and Caracara plancus. Most species were considered rare in the area and not dependent of forest vegetation. -

Birding Tour to Ghana Specializing on Upper Guinea Forest 12–26 January 2018
Birding Tour to Ghana Specializing on Upper Guinea Forest 12–26 January 2018 Chocolate-backed Kingfisher, Ankasa Resource Reserve (Dan Casey photo) Participants: Jim Brown (Missoula, MT) Dan Casey (Billings and Somers, MT) Steve Feiner (Portland, OR) Bob & Carolyn Jones (Billings, MT) Diane Kook (Bend, OR) Judy Meredith (Bend, OR) Leaders: Paul Mensah, Jackson Owusu, & Jeff Marks Prepared by Jeff Marks Executive Director, Montana Bird Advocacy Birding Ghana, Montana Bird Advocacy, January 2018, Page 1 Tour Summary Our trip spanned latitudes from about 5° to 9.5°N and longitudes from about 3°W to the prime meridian. Weather was characterized by high cloud cover and haze, in part from Harmattan winds that blow from the northeast and carry particulates from the Sahara Desert. Temperatures were relatively pleasant as a result, and precipitation was almost nonexistent. Everyone stayed healthy, the AC on the bus functioned perfectly, the tropical fruits (i.e., bananas, mangos, papayas, and pineapples) that Paul and Jackson obtained from roadside sellers were exquisite and perfectly ripe, the meals and lodgings were passable, and the jokes from Jeff tolerable, for the most part. We detected 380 species of birds, including some that were heard but not seen. We did especially well with kingfishers, bee-eaters, greenbuls, and sunbirds. We observed 28 species of diurnal raptors, which is not a large number for this part of the world, but everyone was happy with the wonderful looks we obtained of species such as African Harrier-Hawk, African Cuckoo-Hawk, Hooded Vulture, White-headed Vulture, Bat Hawk (pair at nest!), Long-tailed Hawk, Red-chested Goshawk, Grasshopper Buzzard, African Hobby, and Lanner Falcon. -

Visual Adaptations of Diurnal and Nocturnal Raptors
Seminars in Cell and Developmental Biology 106 (2020) 116–126 Contents lists available at ScienceDirect Seminars in Cell & Developmental Biology journal homepage: www.elsevier.com/locate/semcdb Review Visual adaptations of diurnal and nocturnal raptors T Simon Potiera, Mindaugas Mitkusb, Almut Kelbera,* a Lund Vision Group, Department of Biology, Lund University, Sölvegatan 34, S-22362 Lund, Sweden b Institute of Biosciences, Life Sciences Center, Vilnius University, Saulėtekio Av 7, LT-10257 Vilnius, Lithuania HIGHLIGHTS • Raptors have large eyes allowing for high absolute sensitivity in nocturnal and high acuity in diurnal species. • Diurnal hunters have a deep central and a shallow temporal fovea, scavengers only a central and owls only a temporal fovea. • The spatial resolution of some large raptor species is the highest known among animals, but differs highly among species. • Visual fields of raptors reflect foraging strategies and depend on the divergence of optical axes and on headstructures • More comparative studies on raptor retinae (preferably with non-invasive methods) and on visual pathways are desirable. ARTICLE INFO ABSTRACT Keywords: Raptors have always fascinated mankind, owls for their highly sensitive vision, and eagles for their high visual Pecten acuity. We summarize what is presently known about the eyes as well as the visual abilities of these birds, and Fovea point out knowledge gaps. We discuss visual fields, eye movements, accommodation, ocular media transmit- Resolution tance, spectral sensitivity, retinal anatomy and what is known about visual pathways. The specific adaptations of Sensitivity owls to dim-light vision include large corneal diameters compared to axial (and focal) length, a rod-dominated Visual field retina and low spatial and temporal resolution of vision. -

Uncorrected Proof
Policy and Practice UNCORRECTED PROOF BBarney_C011.inddarney_C011.indd 117979 99/13/2008/13/2008 44:11:24:11:24 PPMM UNCORRECTED PROOF BBarney_C011.inddarney_C011.indd 118080 99/13/2008/13/2008 44:11:24:11:24 PPMM Copy edited by Richard Beatty 11 Conservation Values from Falconry Robert E. Kenward Anatrack Ltd and IUCN-SSC European Sustainable Use Specialist Group, Wareham, UK Introduction Falconry is a type of recreational hunting. This chapter considers the conser- vation issues surrounding this practice. It provides a historical background and then discusses how falconry’s role in conservation has developed and how it could grow in the future. Falconry, as defi ned by the International Association for Falconry and Conservation of Birds of Prey (IAF), is the hunting art of taking quarry in its natural state and habitat with birds of prey. Species commonly used for hunt- ing include eagles of the genera Aquila and Hieraëtus, other ‘broad-winged’ members of the Accipitrinae including the more aggressive buzzards and their relatives, ‘short-winged’ hawks of the genus Accipiter and ‘long-winged’ falcons (genus Falco). Falconers occur in more than 60 countries worldwide, mostly in North America, the Middle East, Europe, Central Asia, Japan and southern Africa. Of these countries, 48 are members of the IAF. In the European Union falconry Recreational Hunting, Conservation and Rural Livelihoods: Science and Practice, 1st edition. Edited by B. Dickson, J. Hutton and B. Adams. © 2009 Blackwell Publishing, UNCORRECTEDISBN 978-1-4051-6785-7 (pb) and 978-1-4051-9142-5 (hb). PROOF BBarney_C011.inddarney_C011.indd 118181 99/13/2008/13/2008 44:11:24:11:24 PPMM Copy edited by Richard Beatty 182 ROBERT E. -
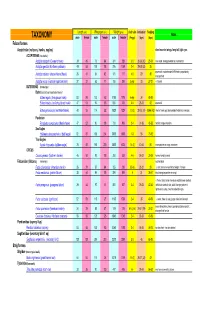
Avian Taxonomy
Length (cm) Wing span (cm) Weight (gms) cluch size incubation fledging Notes TAXONOMY male female male female male female (# eggs) (days) (days) Falconiformes Accipitridae (vultures, hawks, eagles) short rounded wings; long tail; light eyes ACCIPITRINAE (true hawks) Accipiter cooperii (Cooper's hawk) 39 45 73 84 341 528 3-5 30-36 (30) 25-34 crow sized; strongly banded tail; rounded tail Accipiter gentilis (Northern goshawk) 49 58 101 108 816 1059 2-4 28-38 (33) 35 square tail; most abundant NAM hawk; proportionaly 26 31 54 62 101 177 4-5 29 30 Accipiter striatus (sharp-shinned hawk) strongest foot Accipiter nisus (Eurasian sparrowhawk ) 37 37 62 77 150 290 5-Apr 33 27-31 Musket BUTEONINAE (broadwings) Buteo (buzzards or broad tailed hawks) Buteo regalis (ferruginous hawk) 53 59 132 143 1180 1578 6-Apr 34 45-50 Buteo lineatus (red-shouldered hawk) 47 53 96 105 550 700 3-4 28-33 42 square tail Buteo jamaicensis (red-tailed hawk) 48 55 114 122 1028 1224 1-3 (3) 28-35 (34) 42-46 (42) (Harlan' hawk spp); dark patagial featehres: immature; Parabuteo Parabuteo cuncincutus (Harris hawk) 47 52 90 108 710 890 2-4 31-36 45-50 reddish orange shoulders Sea Eagles Haliaeetus leucocephalus (bald eagle) 82 87 185 244 3000 6300 1-3 35 70-92 True Eagles Aquila chrysaetos (golden eagle) 78 82 185 220 3000 6125 1-4 (2) 40-45 50 white patches on wings: immature; CIRCUS Circus cyaneus (Northern harrier) 46 50 93 108 350 530 4-6 26-32 30-35 hovers; hunts by sound Falconidae (falcons) (longwings) notched beak Falco columbarius (American merlin) 26 29 57 64