Automatic Brain Extraction from MRI of Human Head Scans Using Fuzzy Logic and Bridge Building Algorithm
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

C O N T E N T S
22.12.2014 1 C O N T E N T S Sixteenth Series, Vol.VI, Third Session, 2014/1936 (Saka) No.21, Monday, December 22, 2014/Pausa 1, 1936 (Saka) S U B J E C T P A G E S OBITUARY REFERENCE 2 ORAL ANSWERS TO QUESTIONS Starred Question No.401 to 405 4-44 WRITTEN ANSWERS TO QUESTIONS Starred Question Nos.406 to 420 45-98 Unstarred Question Nos.4601to 4830 99-495 The sign + marked above the name of a Member indicates that the Question was actually asked on the floor of the House by that Member. 22.12.2014 2 PAPERS LAID ON THE TABLE 496-508 MESSAGE FROM RAJYA SABHA 509 BUSINESS ADVISORY COMMITTEE 509 10th Report COMMITTEE ON WELFARE OF SCHEDULED 509 CASTES AND SCHEDULED TRIBES Study Tour Report COMMITTEE ON PAPERS LAID ON THE TABLE 510 1st and 2nd Reports STANDING COMMITTEE ON AGRICULTURE 510 5th Report STANDING COMMITTEE ON INFORMATION 510-511 TECHNOLOGY 1st to 4th Reports STANDING COMMITTEE ON DEFENCE 511 2nd to 5th Reports STANDING COMMITTEE ON ENERGY 511-512 st rd 1 to 3 Reports STANDING COMMITTEE ON EXTERNAL AFFAIRS 512 3rd and 4th Reports STANDING COMMITTEE ON FINANCE 512 4th, 8th, and 9th Reports STANDING COMMITTEE ON FOOD, CONSUMER 513 AFFAIRS AND PUBLIC DISTRIBUTION 1st and 2nd Reports STANDING COMMITTEE ON LABOUR 513 3rd Report 22.12.2014 3 STANDING COMMITTEE ON COAL AND STEEL 513-514 1st to 6th Reports STANDING COMMITTEE ON COMMERCE 514 115th and 116th Reports STANDING COMMITTEE ON HOME AFFAIRS 515 182nd and 183rd Reports STATEMENTS BY MINISTERS (i)Status of implementation of the recommendations contained in the 251st Report of the Standing Committee on Industry on ‘functioning of Prime Minister’s Employment Generation Programme (PMEGP)’, pertaining to the Ministry of Micro, Small and Medium Enterprises. -

THE TAMIL NADU Dr. M.G.R. Medical University, Chennai
THE TAMIL NADU Dr. M.G.R. Medical University, Chennai M.Ch. Affiliated Colleges/Institutions for the Academic Year 2020‐21 GOVERNMENT COLLEGES Sl.No. Inst. Name of the Institution Branch Sanctioned Code Intake 1 006 Chengalpattu Medical College, M.Ch. Plastic & Reconstructive Surgery 4 Chengalpattu – 603 001. M.Ch. Surgical Oncology 2 M.Ch. Urology 2 2 007 Coimbatore Medical College, M.Ch. Neuro-Surgery 4 Avinashi Road, M.Ch. Paediatric Surgery 4 Coimbatore – 641 014. M.Ch. Plastic & Reconstructive Surgery 2 M.Ch. Surgical Oncology 1 3 010 Government Mohan Kumaramangalam Medical M.Ch. Neuro-Surgery 2 College, Salem – 636 030. M.Ch. Paediatric Surgery 2 M.Ch. Plastic & Reconstructive Surgery 2 M.Ch. Surgical Gastroenterology 2 M.Ch. Urology 4 4 087 Kanyakumari Government Medical College, M.Ch. Neuro-Surgery 2 Kanyakumari 5 004 Kilpauk Medical College, M.Ch. Plastic & Reconstructive Surgery 6 Kilpauk, Chennai 600010 M.Ch. Surgical Gastroenterology 2 M.Ch. Surgical Oncology 6 M.Ch. Urology 6 6 001 Madras Medical College, M.Ch. Cardio Vascular & Thoracic Surgery 14 E.V.R. Periyar Salai, M.Ch. Endocrine Surgery 2 Park Town, M.Ch. Neuro-Surgery 9 Chennai - 600 003 M.Ch. Paediatric Surgery 9 M.Ch. Plastic & Reconstructive Surgery 8 M.Ch. Surgical Gastroenterology 6 M.Ch. Surgical Oncology 2 M.Ch. Urology 8 M.Ch. Vascular Surgery 6 7 003 Madurai Medical College, M.Ch. Cardio Vascular & Thoracic Surgery 2 Madurai-625 020 . M.Ch. Neuro-Surgery 4 M.Ch. Paediatric Surgery 2 M.Ch. Plastic & Reconstructive Surgery 4 M.Ch. Surgical Gastroenterology 3 M.Ch. -

Institute Name - Thiagarajar College, Madurai -625009 India Rankings 2019 ID - IR-C-C-36513 / IR-O-C-36513
Institute Name - Thiagarajar College, Madurai -625009 India Rankings 2019 ID - IR-C-C-36513 / IR-O-C-36513 Parameter Students opting for higher studies (UG - PG / Others) Graduating Number of S.No. year of the Name of the University/Institutions Students Year of admission 3A.GPHE Student admitted 1 Indian Institute of Technology, Mumbai 1 2 Thiagarajar College, Madurai 85 3 Thiagarajar College of Preceptors , Madurai 27 Thiagarajar College of Engineering , 4 Thiruparankundram 6 5 Thiagarajar School of Management , Madurai 3 6 American College , Madurai 35 7 Lady Doak College 15 8 Alagappa university , Karaikudi 12 9 Alagappa Chettiyar Engineering College 2 10 Annamalai university, Chithambaram 5 11 Madurai Kamarajar University 49 12 Madura College 13 13 Madras University 1 14 Madurai Institute of Social Sciences 6 15 KLN College of Education 12 16 A.K.R. Sourashtra B.Ed College , Madurai 3 17 Annai Fatima college , Thirumangalam 1 Annai Theresa Institute Of Hotel Management & 18 Paramedicals, Palanganatham, 1 19 Anugraha College , Dindigul 1 20 Apollo Institute ,Madurai 1 Arasan Ganeshan College of Education , Sivagasi 21 1 1 2017-18 2018-19 22 Arulanandar College, Karumathut 3 Arumugam Nallamani College of Education , 23 Thiruppalai 3 Arumugam Pillai Seethai Ammal College of Education 24 , Thirupathur 1 25 Asifa College of Education 1 26 Ayira Vaisya College of Education 1 27 Ayya Nadar Janaki Ammal College 1 28 Bharath College of Education , Thanjavur 1 29 Bharathidasan University , Trichy 6 30 Bharathiyar University 3 31 Bishop Heber -

Dr. K. Ruckmani
Dr.K.Ruckmani Tel: +91−98424 84568 ; email: [email protected] Professor & Head Dept of Pharmaceutical Technology Director, Centre for Nanobio Translational REsearch(Autonomous) Bharathidasan Institute of Technology Anna University, Tiruchirappalli 620 024 RESEARCH INTEREST Nanopharmaceuticals Multifunctional smart nanoparticle systems Inhalation Toxicology Impact of Environmental and societal factors in pulmonary diseases Bioactive peptides from natural sources and stabilization EDUCATIONAL QUALIFICATIONS Chartered Management Institute (CMI) Level 5 Certificate Management and Leadership (QCF) Ph.D. (Pharmacy), Department of Pharmaceutical Technology, Jadavpur University, Kolkatta. First lady from Tamil Nadu to be awarded a Doctorate in Pharmacy (2000) M.Pharmacy, Department of Pharmaceutical Technology, Jadavpur University, Kolkatta. University Fifth rank(1994) B.Pharmacy,Madurai Medical College, Madurai Kamarajar University, Madurai. University First Rank(1991) D.Pharmacy ,Thanjavur Medical College, Thanjavur .First Class (1987) NATIONAL LEVEL TEST Qualified in GRATUATE APTITUDE TEST IN ENGINEERING (GATE)-1992 POST DOCTORAL RESEARCH AND FELLOWSHIPS RECEIVED March 2007 - March 2008 BOYSCAST (Better Opportunities for Young Scientists in Chosen Areas of Science & Technology) Fellowship by Department of Science & Technology, Ministry of Science & Technology, Government of India for conducting advanced research / undergoing specialized training in the area of Life Sciences/Drug Delivery systems for a duration of 12 months at -

Tamil Nadu MBBS Cutoff for 85% State Quota Seats
Tamil Nadu MBBS cutoff for 85% State Quota seats Name of College Miscellaneou Open OBC SC ST s (Quota) Rank Scor Rank Scor Rank Scor Rank Scor e e e e Annapoorna Medical Government -- -- 3772 435 1155 325 1724 262 College and Quota 0 8 Hospital, Salem Management 6774 384 -- -- -- -- -- -- Quota Chengalpattu Government 871 525 1102 512 5306 407 1152 326 Medical College, Quota 2 Chengalpattu Christian Medical Management 3419 439 -- -- -- -- -- -- College, Vellore Quota (Institutional (Institutional Quota) Quota) CMC Vellore - (All Management 13730 269 -- -- -- -- -- -- categories) Quota CMC Vellore - Management 2501 465 -- -- -- -- -- -- General Quota Coimbatore Medical Government 464 556 672 536 5681 401 1011 341 College, Coimbatore Quota 8 Dhanalakshmi Government 2505 463 3364 444 1137 327 2128 218 Srinivasan Medical Quota 1 4 College and Management 5427 393 -- -- -- -- -- -- Hospital, Quota Perambalur Dharmapuri Medical Government -- -- 1671 488 7368 376 1289 310 College, Dharmapuri Quota 6 ESIC Medical Government 1195 508 2248 470 8320 363 1568 280 College, K.K. Nagar, Quota 3 Chennai Government Government 938 520 1215 507 6263 392 7544 373 K.A.P.V. Medical Quota College, Trichy Government Karur Government -- -- 2246 470 8536 360 1352 303 Medical College, Quota 2 Karur Government Medical Government 761 531 990 518 5092 411 1122 329 College, Omandurar Quota 4 Estate, Chennai Government Mohan Government 680 536 1108 512 5582 403 9301 351 Kumarmangalam Quota Medical College, Salem Government Medical Government -- -- 2238 470 8469 361 -

Commencement1976.Pdf (4.717Mb)
1976 Digitized by the Internet Archive in 2012 with funding from LYRASIS Members and Sloan Foundation http://archive.org/details/commencement1976 ORDER OF PROCESSION MARSHALS STANLEY CORRSIN JOHN W. GRYDER MATTHEW A. CRENSON WILLIAM H. HUGGINS ELAINE C. DAVIS ROBERT A. LYSTAD HANS GOEDICKE EVANGELOS N. MOUDRIANAKIS ARCHIE GOLDEN EVERETT SCHILLER GERALD S. GOTTERER JOHN P. YOUNG THE GRADUATES MARSHALS ROBERT B. POND OREST RANUM THE DEANS MEMBERS OF THE SOCIETY OF SCHOLARS OFFICERS OF THE UNIVERSITY THE TRUSTEES * MARSHALS BROWN L. MURR FRANCIS E. ROURKE THE FACULTIES * CHIEF MARSHAL RICHARD A. MACKSEY THE CHAPLAINS THE RECIPIENT OF THE MILTON STOVER EISENHOWER MEDAL FOR DISTINGUISHED SERVICE THE PRESENTOR OF THE RECIPIENT OF THE MILTON STOVER EISENHOWER MEDAL FOR DISTINGUISHED SERVICE THE HONORARY DEGREE CANDIDATES THE PROVOST OF THE UNIVERSITY THE PRESIDENT EMERITUS OF THE UNIVERSITY THE CHAIRMAN OF THE BOARD OF TRUSTEES THE PRESIDENT OF THE UNIVERSITY ORDER OF EVENTS STEVEN MULLER President of the University, presiding * * * FANFARE PROCESSIONAL The audience is requested to stand as the Academic Procession moves into the area and to remain standing after the Invocation. " " Earle of Oxford's Marche William Byrd The Peabody Wind Ensemble Richard Higgins, Director * INVOCATION REV. CHESTER WICKWIRE Chaplain, The Johns Hopkins University THE NATIONAL ANTHEM GREETINGS ROBERT D. H. HARVEY Chairman of the Board of Trustees PRESENTATION OF THE RECIPIENT FOR THE MILTON STOVER EISENHOWER MEDAL FOR DISTINGUISHED SERVICE HELEN B. TAUSSIG PRESENTED BY RICHARD S. ROSS Vice President for the Health Divisions and Dean, School of Medicine * PRESENTATION OF NEW MEMBERS OF THE SOCIETY OF SCHOLARS LEROY E. -

Directorate of Medical Education
1 Directorate of Medical Education Hand Book on Right to Information Act - 2005 2 CHAPT ER DETAILS PAGE NO NO. 1. INTRODUCTION 3 2 PARTICULARS OF ORGANISATION, FUNCTIONS AND DUTIES 5 3 POWERS AND DUTIES OF OFFICERS AND EMPLOYEES 80 4 RULES, REGULATIONS,INSTRUCTIONS, MANUAL AND RECORDS 91 FOR DISCHARGING FUNCTIONS 5 PARTICULARS OF ANY ARRANGEMENT THAT EXISTS FOR 93 CONSULTATION WITH OR REPRESENTATION BY THE MEMBERS OF THE PUBLIC IN RELATION TO THE FORMULATION OF ITS POLICY OR IMPLEMENTATION THEREOF 6 A STATEMENT OF THE CATEGORIES OF DOCUMENTS THAT ARE 94 HELD BY IT OR UNDER ITS CONTROL 7 A STATEMENT OF BOARDS, COUNCIL, COMMITTEES AND OTHER 95 BODIES CONSTITUTED AS ITS PART 8 THE NAMES, DESIGNATION AND OTHER PARTICULARS OF THE 96 PUBLIC INFORMATION OFFICER 9 PROCEDURE FOLLOWED IN DECISION MAKING PROCESS 107 10 DIRECTORY OF OFFICERS AND EMPLOYEES 108 11 THE MONTHLY REMUNERATION RECEIVED BY EACH OF ITS 110 OFFICERS AND EMPLOYEES INCLUDING THE SYSTEM OF COMPENSATION AS PROVIDED IN REGULATIONS. 12 THE BUDGET ALLOCATED TO EACH AGENCY/OFFICERS 111 (PARTICULARS OF ALL PLANS, PROPOSED EXPENDITURES AND REPORTS ON DISBURSEMENT MADE 116 13 THE MANNER OF EXECUTION OR SUBSIDY PROGRAMME 14 PARTICULARS OF RECEIPIENTS OF CONCESSIONS, PERMITS OR 117 AUTHORISATION GRANTED BY IT 15 NORMS SET BY IT FOR THE DISCHARGE OF ITS FUNCTIONS 118 16 INFORMATION AVAILABLE IN AN ELECTRONIC FORM 119 17 PARTICULARS OF THE FACILITIES AVAILABLE TO CITIZENS FOR 120 OBTAINING THE INFORMATION 18 OTHER USEFUL INFORMATION 121 3 CHAPTER-1 - INTRODUCTION 1.1This handbook is brought out by the Directorate of Medical Education (Government of Tamil Nadu) , Chennai as required by the Right to Information Act, 2005. -
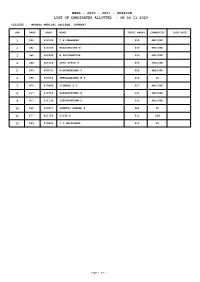
Mbbs - 2020 - 2021 - Session List of Candidates Allotted - on 30.11.2020
MBBS - 2020 - 2021 - SESSION LIST OF CANDIDATES ALLOTTED - ON 30.11.2020 COLLEGE : MADRAS MEDICAL COLLEGE, CHENNAI. SNO RANK ARNO NAME TOTAL MARKS COMMUNITY JOIN DATE 1 366 612545 T N VARSHNEE 630 MBC/DNC 2 382 616049 KALAISELVAN M 630 MBC/DNC 3 385 603999 M PRIYAMAYURA 629 MBC/DNC 4 388 605468 JEYA SURYA S 629 MBC/DNC 5 394 609572 KISHOREKUMAR G 628 MBC/DNC 6 396 600054 DEEPALAKSHMI M P 628 SC 7 400 610886 VIGNESH S V 627 MBC/DNC 8 417 611760 PRADEEPKUMAR K 626 MBC/DNC 9 427 611136 JEEVANANTHAM S 626 MBC/DNC 10 466 619457 GOMATHI SANKAR K 624 SC 11 677 622154 DIVYA D 612 SCA 12 689 619840 C V MELESHNAA 612 SC Page 1 of 1 MBBS - 2020 - 2021 - SESSION LIST OF CANDIDATES ALLOTTED - ON 30.11.2020 COLLEGE : STANLEY MEDICAL COLLEGE, CHENNAI. SNO RANK ARNO NAME TOTAL MARKS COMMUNITY JOIN DATE 1 367 604825 NAFEELA A 630 BCM 2 371 600327 FADHIL MOHAMMED S 630 BCM 3 378 603632 ABDUL AZIZ H 630 BCM 4 403 623146 ROJA MARIYAM S 627 BCM 5 446 616605 SARAVANA SIDHAARTH S 625 MBC/DNC 6 449 626689 BHUVANESWARI N 625 MBC/DNC 7 453 617019 HARSHATH N J 625 MBC/DNC 8 455 613595 ABARNA S 624 MBC/DNC 9 459 606977 CHEMPARUTHI J V 624 MBC/DNC 10 462 605684 ANANDHAKRISHNAN M 624 MBC/DNC 11 464 619302 KAVI SURYA P N 624 BCM 12 465 603930 SRIKUMARAN P 624 MBC/DNC 13 474 619127 SARAVANAN P 623 MBC/DNC 14 477 614245 DHIWAKAR R S 622 MBC/DNC 15 495 619285 RUBHADEVI A P 621 MBC/DNC 16 496 607480 NIVETHASHRI S 621 MBC/DNC 17 507 607244 PRITHIVINATHAN A 620 MBC/DNC 18 522 613833 DHANASEKAR S 620 MBC/DNC 19 530 613356 PRIYANKA M 620 MBC/DNC 20 533 617782 KAAVIYAA V G 619 MBC/DNC 21 557 619757 SHAKTHI SRI G 618 MBC/DNC 22 568 606916 PRIYADHARSHINI A 617 MBC/DNC 23 569 623529 MAHARAJAN M 617 MBC/DNC 24 580 610564 SIVASUTHAN V 617 MBC/DNC 25 581 622201 LOKESH A 617 MBC/DNC 26 582 608892 SOUNDARAMOULI S 617 MBC/DNC 27 584 621030 G SANJAY 617 MBC/DNC 28 585 607446 LAKSHMAN R L 617 MBC/DNC 29 586 617527 ROOPINI R 617 MBC/DNC 30 593 606670 VIGNESH S 616 MBC/DNC Page 1 of 2 MBBS - 2020 - 2021 - SESSION LIST OF CANDIDATES ALLOTTED - ON 30.11.2020 COLLEGE : STANLEY MEDICAL COLLEGE, CHENNAI. -

Patron-In-Chief Honorary Fellows Founder Fellows
Patron-in-Chief 1. Dr. Rajendra Prasad 2. Dr. S. Radhakrishnan 3. Dr. Zakir Husain 4. Shri V.V. Giri 5. Shri Fakhruddin Ali Ahmed 6. Shri Giani Zail Singh Honorary Fellows 1. Shri Jawahar Lal Nehru 2. Dr. B.C. Roy 3. Major Genl. S.L. Bhatia 4. Col. R.N. Chopra 5. Dr. H.M. Lazarus 6. Dr. Jivaraj N. Mehta 7. Dr. A. Lakshamanswami Mudaliar 8. Dr. N.A. Purandare 9. Major Genl. S.S. Sokey 10. Dr. A.C. Ukil 11. Dr. Sushila Nayar 12. Smt. Indira Gandhi 13. Dr. V.T.H. Gunaratne 14. Dr. Dharmendra 15. Shri P.V. Narasimha Rao Founder Fellows 1. Dr. Madan Lal Aggarwal 2. Dr. B.K. Aikat 3. Dr. S.T. Achar 4. Dr. (Col.) Amir Chand 5. Dr. A.A. Ayer 6. Dr. Santokh Singh Anand 7. Dr. R.B. Arora 8. Dr. L.H. Athle 9. Dr. A.V. Baliga 10. Dr. Baldev Singh 11. Dr. Bankat Chandra 12. Dr. A.K. Basu 13. Dr. B.B. Bhatia 14. Dr. T.N. Banerjee 15. Dr. Bimal Chandra Bose 16. Dr. J.C. Banerjee 17. Dr. E.J. Borges 18. Dr. P.V. Cherian 19. Dr. R.N. Chaudhuri 20. Dr. G. Coelho 21. Dr. R.A.F. Cooper 22. Dr. (Lt.Genl.) D.N. Chakravarti 23. Dr. L.W. Chacko 24. Dr. M.K.K. Menon 25. Dr. Subodh Mitra 26. Dr. (Capt) P.B. Mukherjee 27. Dr. S.R. Mukherjee 28. Dr. B. Mukhopadhaya 29. Dr. M. Naidu 30. Dr. B. Narayana 31. Dr. C.G. -
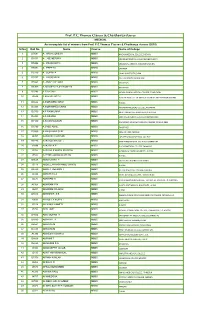
Prof. P.C.Thomas Classes & Chaithanya Classes
Prof. P.C.Thomas Classes & Chaithanya classes MEDICAL An incomplete list of winners from Prof. P.C.Thomas Classes & Chaithanya classes (2019) Sl.No Roll No Name Course Name of College 1 D5691 A . ARUN LOKESH MBBS MADHA MEDICAL COLLEGE,CHENNAI 2 D6553 A . JOE MERWIN MBBS SRI VENKATESWARA COLLEGE,PONDICHERRY 3 D5446 A . PRASHANTH MBBS VELAMMAL MEDICAL COLLEGE,MADURAI 4 D5882 A . PREETHI MBBS UKRAINE 5 D6119 A . SUPRIYA MBBS JINAN UNIVERSITY,CHINA 6 D5737 A . VASIGARAN MBBS ISM UNIVERSITY,BANGALURU 7 D5863 A .ANGELINFLORA MBBS PHILIPINES 8 D6909 A AHAMED VAJEERUDEEN MBBS PHILIPINES 9 D0743 A B BARANI MBBS INDIRA GANDHI MEDICAL COLLEGE,PONDICHERY 10 J5849 A M HASIL BECK MBBS AZEEZIA INSTITUTE OF MEDICAL SCIENCES MEEYYANOOR KOLLAM 11 D6522 A MOHAMED ISHAK MBBS RUSSIA 12 D7203 A MOHAMMED UNAIS MBBS ANDAMAN MEDICAL COLLEGE,ANDAMAN 13 D0750 A P RAMKUMAR MBBS NORTH PHILIPINES UNIVERSITY,PHILIPINES 14 D0494 A S ASHISH MBBS SREE MOOKAMBIKA COLLEGE,KANYAKUMARI 15 D7180 A SIVASHAAGARI MBBS MANAKULA VINAYAKA MEDICAL COLLEGE,PONDICHERRY 16 D6790 A SYED ADHIL MBBS PHILIPINES 17 D0066 A.VAISHNAVI SHRI MBBS SRM COLLEGE,CHENNAI 18 J4957 AADARSH HARIDAS MBBS JUBILEE MISSION MEDICAL COLLEGE 19 D7275 AAKAASH RAHUL J MBBS KARPAKAM MEDICAL COLLEGE,COIMBATORE 20 A0199 AAKASH K P MBBS P K DAS MEDICAL COLLEGE PALAKKAD 21 J5326 AASHNA P MURALIDHARAN MBBS MORDOVIA STATE UNIVERSITY , RUSSIA 22 J5532 AATHIRA OMANAKUTTAN MBBS RUSSIA 23 D5628 ABARAJITHA T MBBS UV GULAS UNIVERSITY,PHILIPINES 24 J5118 ABDULLAH MOHAMED UVAISE MBBS RUSSIA 25 D6229 ABDUR REHMAN T MBBS KIL PAUK MEDICAL COLLEGE,CHENNAI 26 J5342 ABHIPRIYA A MBBS GOVT. -

Pushpalata Vidya Mandir Alumni Bulletin
PUSHPALATA VIDYA MANDIR ALUMNI BULLETIN I Batch (2013 – 14) SL.NO NAMES PLACEMENT 1 V.S. Seralathan IIT, Guwahati 2 R. Selva Sundari IIM, Indore 3 L. Shanmuga Priya North Carolina State University, USA 4 Sushmitha Prasad MVJ Medical College & Research Hospital, Bangalore. 5 Aparna Sivaraman Prabha Central University of Tamilnadu, Tiruvarur 6 Hathim Syed Mohamed IIT, Madras II Batch (2014 – 15) SL.NO NAMES PLACEMENT 1 H.S. Srividya NIT Trichy Admitted to Stanford University, USA to pursue M.S. 2 N. Delfin Shelty CMC, Vellore 3 Iragaraju Anjani Harika D.S. National Law University, Vizag 4 Gopi.S NIT, Trichy 5 Aazim Bill Se Yaswant IIT, Roorkee 6 G.S. Tharani Ramya CA, Inter III Batch (2015 – 16) SL.NO NAMES PLACEMENT 1 K. Adharsh Rishi Kiran IIT, Madras 2 D. Lavanya Ruba Malini Tirunelveli Medical College 3 S. Anantharaman BITS Pilani, Hyderabad 4 L. Wellington Daniel IIM, Indore 5 Z. Zahi Kamal Tamilnadu Dr. Ambedkar Law University IV Batch (2016 – 17) SL.NO NAMES PLACEMENT 1 Vignesh C. Arun NIT, Trichy 2 V. Sanghamithra NIT, Trichy 3 T.M. Snekaa Stanley Medical College, Chennai 4 Basheer Ashik. P Anna University 5 S. Rajesh School of Planning & Architecture, Vijayawada 6 V. Harini CA Inter 7 R. Jothi Vikranth Indian Institute of Information Technology Design and Manufacturing, Kancheepuram 8 S.S. Jaya Dharshana LBS Centre for Science & Tech, Thiruvananthapuram 9 S. Sakthi Ram Thoothukudi Medical College 10 K.M. Aakash ESIC Medical College, Chennai 11 A.P. Saraswathi CA Inter 12 N. Alsia Fathima SRM Chennai Medical College Hospital & Research Centre 13 R. -

1 Government of Tamilnadu Prospectus for Admission
GOVERNMENT OF TAMILNADU PROSPECTUS FOR ADMISSION TO MBBS / BDS DEGREE COURSES IN TAMILNADU GOVERNMENT MEDICAL / DENTAL COLLEGES, GOVERNMENT ERODE MEDICAL COLLEGE AND HOSPITAL, PERUNDURAI, ERODE AND GOVT. SEATS IN SELF FINANCING MEDICAL / DENTAL COLLEGES AFFILIATED TO THE TAMILNADU DR.M.G.R. MEDICAL UNIVERSITY & RAJAH MUTHIAH MEDICAL / DENTAL COLLEGE AFFILIATED TO ANNAMALAI UNIVERSITY, CHIDHAMBARAM. ESIC MEDICAL COLLEGE AND PGIMSR, K.K.NAGAR, CHENNAI. 2020-2021 SESSION As per G.O (D) No. 06, Health and Family Welfare Dept., dated 03.11.2020 and as amended from time to time). Last date for submission of online application form 12 .11.2020 upto 5.00 P.M Website : www.tnhealth.tn.gov.in www.tnmedicalselection.org Cost 500/- 1 IMPORTANT DATES: (Tentative) 1 Date of Notification 03.11.2020 2 Date of Commencement of online application 03.11.2020 3 Last date for online submission of application 12.11.2020 upto 5:00 P.M. 4 Address to which the filled-in online application THE SECRETARY, along with enclosures are to be sent. SELECTION COMMITTEE, (for Special Category candidates) DIRECTORATE OF MEDICAL EDUCATION, 162, PERIYAR E.V.R. HIGH ROAD, KILPAUK, CHENNAI – 600 010. 5 Tentative date of declaration of Rank list 16.11.2020 6 Details of counselling Will be hosted in the official websites. 7 Commencement of courses 15.12.2020 8 Closure of admission 31.12.2020 9 Official websites www.tnhealth.tn.gov.in www.tnmedicalselection.org 10 Helpline Number 1. 044-28364822 2. 9884224648 3. 9884224649 4. 9884224745 5. 9884224746 2 IMPORTANT INFORMATION: 1) The online submission of application form for admission to MBBS/BDS Degree Courses 2020-2021 session in Tamil Nadu Government Medical / Dental Colleges, Government Erode Medical College and Hospital, Perundurai, Erode, Government seats in Self- Financing Medical / Dental Colleges affiliated to the Tamil Nadu Dr.M.G.R.