Curriculum Vitae of Srinivas Aluru
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TWAS an Rep IMP
annual re2p0or1t 0 TWAS, the academy of sciences for the developing world, is an autonomous international organization that promotes scientific capacity and excellence in the South. Founded in 1983 by a group of eminent scientists under the leadership of the late Nobel laureate Abdus Salam of Pakistan, TWAS was officially launched in Trieste, Italy, in 1985, by the secretary-general of the United Nations. TWAS has nearly 1,000 members from over 90 countries. More than 80% of its members are from developing countries. A 13-member council directs the Academy activities. A secretariat, headed by an executive director, coordinates the programmes. The Academy’s secretariat is located on the premises of the Abdus Salam International Centre for Theoretical Physics (ICTP) in Trieste, Italy. TWAS’s administration and finances are overseen by the United Nations Educational, Scientific and Cultural Organization (UNESCO) in accordance with an agreement signed by the two organizations. The Italian government provides a major portion of the Academy’s funding. The main objectives of TWAS are to: • Recognize, support and promote excellence in scientific research in the developing world; • Respond to the needs of young researchers in science and technology- lagging developing countries; • Promote South-South and South-North cooperation in science, technology and innovation; • Encourage scientific research and sharing of experiences in solving major problems facing developing countries. To help achieve these objectives, TWAS collaborates with a number of organizations, most notably UNESCO, ICTP and the International Centre for Biotechnology and Genetic Engineering (ICGEB). academy of sciences for the developing world TWAS COUNCIL President Jacob Palis (Brazil) Immediate Past President C.N.R. -

Chapter 29 SCIENCE and TECHNOLOGY
Chapter 29 SCIENCE AND TECHNOLOGY Introduction The major objectives and thrust in Science & Technology (S&T) sector during the Ninth Five Year Plan, inter alia, has been on (a) optimum harnessing of S&T for societal benefits, (b) developing R&D programmes on mission mode, (c) nurturing of outstanding scientists by offering them facilities comparable with international standards, (d) attracting young scientists for taking science as a career, (e) establishing linkages between the industry and research institutions/ laboratories for development and market technology, (f) developing clean and eco friendly technologies, (g) generating maximum resources for R&D from production and service sector, and (h) creating awareness on technology marketing and intellectual property rights issues. Emphasis has also been laid on science, technology and education including promotion of basic research and excellence, S&T communication and popularisation of science, promotion of international science and technology cooperation. 2. In relation to the Ninth Plan thrust outlined above, there has been significant progress in various areas. Efforts have been continued to maintain a strong science base and develop technological competence. In order to strengthen technological capabilities of the Indian industries, both for meeting the national needs and for proving global competitiveness, a number of new initiatives have been launched through Programme Aimed at Technology Self Reliance (PATSER), Technology Development Board (TDB) and Home Grown Technologies. Individual -
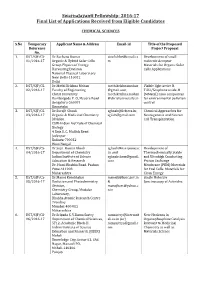
Swarnajayanti Fellowship- 2016-17 Final List of Applications Received from Eligible Candidates
SwarnaJayanti Fellowship- 2016-17 Final List of Applications Received from Eligible Candidates CHEMICAL SCIENCES S.No Temporary Applicant Name & Address Email -id Title of the Proposed Reference Project Proposal No. 1. DST/SJF/CS - Dr.Rachana Kuma r [email protected] Development of small 01/2016-17 Organic & Hybrid Solar Cells m molecule Acceptor Group Physics of Energy Materials for Organic Solar Harvesting Division Cells Applications National Physical Laboratory New Delhi-110012 Delhi 2. DST/SJF/CS - Dr.Mothi Krishna Mohan mothikrishnamohan Visible light active N - 02/2016-17 Faculty of Engineering @gmail.com, TiO2/Graphene oxide-M Christ University mothikrishna.mohan (MMetal) nano composites Kumbalgodu P. O, Mysore Road @christuniversity.in for environmental pollution Bengaluru-560074 control Karnataka 3. DST/SJF/CS - Dr.Surajit Ghosh [email protected]; Chemical Approaches for 03/2016-17 Organic & Medicinal Chemistry [email protected] Neurogenesis and Neuron Division Cell Transplantation CSIR-Indian Institute of Chemical Biology 4 Raja S. C. Mullick Road Jadavpur Kolkata-700032 West Bengal 4. DST/SJF/CS - Dr.Sujit Kumar Ghosh sghosh@iise rpune.ac Development of 04/2016-17 Department of Chemistry .in and Thermochemically Stable Indian Institute of Science sghoshchem@gmail. and Ultrahigh Conducting Education & Research com Proton Exchange Dr. Homi Bhabha Road, Pashan Membrane (PEM) Materials Pune-411008 for Fuel Cells: Materials for Maharashtra Clean Energy 5. DST/SJF/CS - Dr.Manoj Kumbhakar [email protected] Single Molecule 05/2016-17 Radiation and Photochemistry & Spectroscopy of Actinides Division, [email protected] Chemistry Group, Modular o.in Laboratory, Bhabha Atomic Research Centre Trombay Mumbai-400 085 Maharashtra 6. -

CURRICULUM VITAE NAME Jayant B. Udgaonkar DESIGNATION Senior Professor ADDRESS National Centre for Biological Sciences Tata
CURRICULUM VITAE NAME Jayant B. Udgaonkar DESIGNATION Senior Professor ADDRESS National Centre for Biological Sciences Tata Institute of Fundamental Research GKVK Campus Bengaluru 560065 TELEPHONE 91-80-23666150 TELEFAX 91-80-23636662 EMAIL ADRESS [email protected] DATE OF BIRTH March 22, 1960 EDUCATION AND TRAINING DEGREE Years Institution Subjects St. Xaviers College 1976- Major: Chemistry B.Sc. University of Bombay 1979 Minor: Microbiology Mumbai, India Indian Institute Technology, Madras 1979- M.Sc. Department of Chemistry Chemistry 1981 Chennai, India 1981- Cornell University Major: Biochemistry Ph.D. 1986 Section of Biochemistry, Cell and Minor: Applied Physics Molecular Biology Laboratory of Ithaca, NY, U.S.A. Professor GP Hess Post-doctoral Stanford University, Biochemistry 1986- Fellow Department of Biochemistry Laboratory of 1989 Stanford, CA, U.S.A Professor RL Baldwin Research Experience 1990-1995 Reader, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Mumbai and Bangalore 1995-1998 Associate Professor, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore 1998-2007 Professor, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore 2007- Senior Professor, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore Research Interests. Protein folding, dynamics and aggregation; neurodegenerative diseases HONORS AND AWARDS First Rank in the University of Bombay in the B.Sc. examinations in Chemistry (1979) Awarded several University prizes and scholarships, and the National Merit Scholarship by the Government of India. Merit Scholarship, Indian Institute of Technology, Madras (1979 -1981). Fellow of the Jane Coffin Childs Memorial Fund for Medical Research (1986-1989). Biotechnology Career Fellowship, Rockefeller Foundation (1991-1995). -

Swarnajayanti Fellowship- 2016-17 Applications Received from Eligible Candidates
SwarnaJayanti Fellowship- 2016-17 Applications Received from Eligible Candidates (Applicant may inform if there is any mistake or provide missing information to [email protected] & [email protected] before 31 st May, 2017) EARTH & ATMOSPHERIC SCIENCES S.No Temporary Applicant Name & Email -id Title of the Proposed Reference No. Address Project Proposal 1. DST/SJF/E&AS - Dr. Anoop Kumar Mishra [email protected] Developing Near Real 01/2016-17 Centre for Remote sensing Time Flash Flood Risk and Geo-informatics Monitoring Scheme Over Sathyabama University India Jeppiar Nagar, Rajeev Gandhi Road Chennai-600119 Tamilnadu 2. DST/SJF/E&AS - Dr. U. Surendran [email protected] Management of soil and 02/2016-17 Water Management water resources for (Agriculture) Division higher crop productivity Centre for Water in humid tropics of Resources Development Kerala under changing and Management climatic conditions using (CWRDM) crop modeling Kozhikode-673 571 experiments Kerala 3. DST/SJF/E&AS - Dr. Yunus Ali [email protected] -tokyo.ac.jp Estimating future 03/2016-17 Dept. of Geology changes in landslide risk Aligarh Muslim University for Indian and East- Aligarh-202002 Asian nations Uttar Pradesh 4. DST/SJF/E&AS - Dr . Vijayakumar S. Nair [email protected], Stratospheric Black 04/2016-17 Space Physics Laboratory [email protected] Carbon: The unknown Vikram Sarabhai Space climate link Centre Thiruvananthapuram- 695022 Kerala 5. DST/SJF/E&AS - Dr. Suryendu Dutta [email protected] Biosynthetic evolution of 05/2016-17 Department of Earth plant terpenoids on a Sciences rafting continent IIT Bombay Powai Mumbai-400076 Maharashtra S.No Temporary Applicant Name & Email -id Title of the Proposed Reference No. -

Infosys Prize 2016
INFOSYS PRIZE 2016 INFOSYS SCIENCE FOUNDATION “A century ago astronomers, geologists, chemists, physicists, each had an island of his own, separate and distinct from that of every other student of Nature; the whole field of research was then an archipelago of unconnected units. Today all the provinces of study have risen together to form a continent without either a ferry or a bridge.” George Iles From ‘Jottings from a Notebook’, in Canadian Stories (1918) ENGINEERING AND COMPUTER SCIENCE V. KUMARAN Professor, Department of Chemical Engineering, Indian Institute of Science, Bengaluru Prof. V. Kumaran is a senior professor at the Department of Chemical the Indian National Academy of Engineering (2006). More recently, Engineering of the Indian Institute of Science (IISc), Bengaluru. Prof. Kumaran has been elected to the fellowship of the American Prof. Kumaran has been carrying out path-breaking research on Physical Society (2015). many select topics in the areas of fluid mechanics and statistical He has won several prestigious awards including the Shanti Swarup mechanics. Bhatnagar Prize (2000), Swarnajayanti Fellowship (2002), J. C. Bose Kumaran received his B.Tech. in 1987 from IIT-Madras and Ph.D. National Fellowship (2007), the Rustom Choksi Award (2012) for from Cornell University in 1992. He is a fellow of the Indian Academy Excellence in Engineering Research by IISc, the TWAS Prize (2014), of Sciences (1998), Indian National Science Academy (2001), and and the APS-IUSSTF Chair Professorship (2014). The Infosys Prize 2016 in Engineering and Computer Science is awarded to Prof. Viswanathan Kumaran, for his seminal work in complex fluid flows especially in transition and turbulence in soft-walled tubes. -

Database of Scholarship & Fellowship Schemes In
DATABASE OF SCHOLARSHIP & FELLOWSHIP SCHEMES IN SCIENCE & TECHNOLOGY IN INDIA Project sponsored by VIGYAN AUR PRODYOGIKI VIBHAG (NCSTC/RVPSP Division) Department of Science & Technology Government of India *** Database prepared by Prof. Y L Nangia Director Manpower Management Centre New Delhi *** September, 2011 CONTENTS Page No ACKNOWLEDEMENT SCHEMES INDEX i-xii ABOUT THE DATABASE Introduction I Objects II Methodology III S&T Scholarship & Fellowship Schemes in India IV PART - I S & T Scholarship Schemes in India 1-117 PART - II S&T Fellowship Schemes in India 118-225 PART - III S&T Loan Scholarship Schemes in India 226-230 PART - IV S&T Award Schemes in India 231-245 PART - V S&T Associateship/Assistantship Schemes in India 246-251 SCHEMES INDEX In Alphabetic Order PART-I – S&T Scholarship Schemes in India S. No. Page No. 1. Aegis School of Business & Telecommunication 1 Alexander Graham Bell Scholarship 2. BEML Limited, Ministry of Defence 4 BEML Scholarship for SC/ST Students 3. Central Board of Secondary Education 6 CBSE Merit Scholarship Scheme for Professional Course 4. Central Board of Secondary Education 7 Central Sector Scheme of Scholarship for College & University Students 5. Coal India Limited, Ministry of Coal 8 Coal India Scholarship for BPL Students 6. Coal India Limited, Ministry of Coal 10 Coal India Scholarship Scheme for Wards of Land Oustees/ Displaced Persons 7. Delhi College of Engineering 13 DCE Scholarship for M.E./Ph/ D 8. Department for the Welfare of SC/ST/OBC/Minorities 14 Merit Scholarship to SC/ST/OBC/Minorities (including Jain Community) 9. Department of Science and Technology (DST) 15 INSPIRE Programme – Scholarship for Higher Education (SHE) 10. -

DST Selects 2020 Swarnajayanti Fellows a Total of 21 Scientists
DST selects 2020 SwarnaJayanti Fellows A total of 21 scientists associated with innovative research ideas in the areas of life sciences, chemical sciences, mathematics, earth and atmospheric, physical sciences, and engineering and the potential of making an impact on R&D have been selected for the SwarnaJayanti fellowship. Scientists selected for the award will be allowed to pursue unfettered research with freedom and flexibility in terms of expenditure as approved in the research plan. The Swarnajayanti Fellowships‟ scheme was instituted by Govt. of India to commemorate India's fiftieth year of Independence, provides special assistance and support to a selected number of young scientists with a proven track record to enable them to pursue basic research in frontier areas of science and technology. Under the scheme, the awardees are given support by the Department of Science & Technology, GOI, which will cover all the requirements for performing the research and will include a fellowship of Rs. 25,000/- per month for five years. In addition to this, DST supports the awardees by giving them a research grant of 5 lakh Rupees for 5 years. The fellowship is provided in addition to the salary they draw from their parent Institution. In addition to fellowship, grants for equipments, computational facilities, consumables, contingencies, national and international travel, and other special requirements, if any, is covered based on merit. The fellowships are scientists‟ specific and not institution-specific, very selective, and have close academic monitoring. Prof Ashutosh Sharma, Secretary, DST said, “As a new policy measure starting this year, it was decided to support much larger number of young scientists under the SJF in order to recognize, motivate and empower them to work at their highest potential.” A list of 21 Scientists selected for the 2020 fellowship through a rigorous three-layered screening process is given in Annex-I. -

Partha Sarathi Mukherjee
Partha Sarathi Mukherjee Department of Inorganic & Physical Chemistry Indian Institute of Science, Bangalore-560012, Karnataka, India. Tel No: +91-80-2293-3352(O); +91-80-2360-0023(R) Mobile: 09986248948; FAX: +91-80-2360-1552 E-mail: 1) psm@ iisc.ac.in 2) [email protected] ________________________________________________________________________ Present position: Professor, Inorganic & Physical Chemistry Dept., Indian Institute of Science, Bangalore. Research fields: Supramolecular nanomaterials, Organic nanomaterials, Molecular sensors, Catalysis in nanocages. Summary: • 21 years inorganic chemistry research in academia • 16 years teaching experience in chemistry at honours/PG level • Co-author of 180 publications in peer-reviewed journals • Delivered over 200 lectures (invited, plenary, keynote and institute/departmental colloquia) in India and abroad. Educational Qualifications: * 09/1998–01/2002 Doctor of Philosophy (Chemistry), Indian Association for the Cultivation of Science, Kolkata, India. Thesis title: “Synthesis, crystal structure and low temperature magnetic behaviour of Cu(II) polynuclear complexes of amines and their derivatives using different bridging ligands”. Supervisor: Prof. Nirmalendu Ray Chaudhuri * 1996-1998 Master of Science with specialization in Inorganic Chemistry (class 1), Jadavpur University, Kolkata, India. * 1992-1995 Bachelor of Science (Honours in Chemistry, class 1), The University of Burdwan, India. Awards and Fellowships: *2020 Elected Fellow of the Indian Academy of Sciences (FASc) *2018 Member of the Editorial Advisory Board of Inorganica Chimica Acta, a journal published by the Elsevier. *2018 Selected for Deshpande National Award *2016 Shanti Swarup Bhatnagar Prize in Chemical Sciences for the year 2016 *2016 Editorial Advisory Board Member of “Inorganic Chemistry Frontiers”, a journal published by the Royal Society of Chemistry (U. K.) *2016 Associate Editor, Inorganic Chemistry (ACS-Journal). -

Curriculum Vitae Charusita Chakravarty
CURRICULUM VITAE CHARUSITA CHAKRAVARTY Department of Chemistry Indian Institute of Technology-Delhi Hauz Khas, New Delhi 110016 Email: [email protected] Tel: +91 11 2659 1510 Fax: +91 11 2686 2122 EDUCATION October '90 Ph.D in Chemistry Theoretical Chemistry Department, University of Cambridge, UK Thesis Title:Open-shell van der Waals Complexes: Spectra and Dynamics of Ar-OH Research Supervisor: Dr. D. C. Clary June '87 B.A. (Hons.) in Natural Sciences (Chemistry, First Class) Queens' College, University of Cambridge, UK July '85 B.Sc. (Hons.) in Chemistry (First Class) St. Stephens College, University of Delhi, India APPOINTMENTS Aug. '02 - Associate Professor, Department of Chemistry Indian Institute of Technology, New Delhi, India. Oct. '94 - Assistant Professor, Department of Chemistry July '02 Indian Institute of Technology, New Delhi, India. Sept. '93 - Gulbenkian Junior Research Fellowship from Churchill College, Cambridge. Sept. '94 Affiliated to the Department of Chemistry, University of Cambridge, UK Nov. '92 - Department of Physics Aug. '93 Indian Institute of Technology-Delhi, India March '91 - Department of Chemistry, University of California, Santa Barbara, USA Sept. '92 Postdoctoral research associate with Prof. Horia Metiu AWARDS/PRIZES • Fellow of the Indian Academy of Sciences, Bangalore. • Swarnajayanti Fellowship 2003-2004, Department of Science and Technology, New Delhi. • Bronze Medal of the Chemical Research Society of India, 2004. • B. M. Birla Science Award in Chemistry for 1999. • Anil Kumar Bose Memorial Award for 1999, Indian National Science Academy, New Delhi. • Associate Member of the Abdus Salam International Centre for Theoretical Physics, Trieste, Italy (1996-2003). • INSA Medal for Young Scientists 1996, Indian National Science Academy, New Delhi. -

NASI Scopus Young Scientist Award in 2014
NASI SCOPUS th YOUNG 12 SCIENTIST AWARDS 2018 Supporting India’s ambition to become a "Knowledge Superpower" NASI-SCOPUS Young Scientist Awards 2018 About Scopus Scopus is the largest abstract and citation database of peer-reviewed literature: scientific journals, books and conference proceedings. Delivering a comprehensive overview of the world’s research output in the fields of science, technology, medicine, social sciences, and arts and humanities, Scopus features smart tools to track, analyze and visualize research. As research becomes increasingly global, interdisciplinary and collaborative, you can make sure that critical research from around the world is not missed when you choose Scopus. “Speed is very important...I can easily identify what I need to know, read it, digest it, and move on to the next one.” James, Research Pathologist, Medical Device R&D, Scopus user “Scopus informs every phase of the editorial process. I would not want to do this job without it, and I intend to continue using it throughout my career.” William, Professor of Economics, University of Tennessee “...[Scopus] will allow us to analyze a deeper range of research activity from a wider range of institutions than present, including those institutions from emerging economies that account for a growing portion of the world’s research output and which have shown a great hunger for THE’s trusted global performance metrics.” THE Managing Director Trevor Barratt For more information, please visit: http://www.elsevier.com/solutions/scopus 1 NASI - SCOPUS YOUNG SCIENTIST AWARDS 2018 India, with its knowledge base, is all set to become the next scientific super power. -

Annual Report 2019-20
Annual Report 2019-20 Science and Engineering Research Board Science and Engineering Research Board 2019-20 Annual Report From the Secretary's Desk I feel privileged to present the 9th Annual Report of the Science and Engineering Research Board (SERB), illustrating a comprehensive overview of our activities and achievements in the year 2019-2020. In this year, SERB saw the launch of several new R&D programs that offers a wider landscape for Indian researchers to seek outstanding scientific breakthroughs, making impactful contributions to our fundamental understanding and offer disruptive technologies at the cutting edge. New programs such as SERB-SUPRA (Scientific and Useful Profound Research Advancement) and SERB-Exponential Technologies intend to challenge the Indian researchers to evolve transformative concepts based on original and creative research problems, opening up newer opportunities in national S&T landscape and create the right visibility on the global stage. With ever-increasing outreach, we are at the forefront of developing a right ecosystem to crowd-source outstanding research problems, and delivery of equally unique solutions and knowledge creation. SERB is also on the forefront in bringing wider platforms for multiple players in research and development. SERB-FIRE (Fund for Industrial Research Engagement) is a new model of public-private partnership, put in motion in the reporting period, to serve as a new framework for industry-oriented research, along with capital contributions from industries. SERB-STAR (Science and Technology Award for Research) program was instituted to recognize and reward outstanding research performance in frontier areas of science and engineering. It is envisaged that SERB-STAR will be an appropriate recognition for investigators, by acknowledging their exemplary contributions in research and sustain their motivation for to undertake pioneering research.