Roles of DDX5 in the Tumorigenesis, Proliferation, Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Autophagy: from Basic Science to Clinical Application
nature publishing group REVIEW See COMMENTARY page XX Autophagy: from basic science to clinical application J Va n L i m b e r g e n 1 , 2 , 3 , C S t e v e n s 4 , E R N i m m o 1 , D C W i l s o n 2 , 3 a n d J S a t s a n g i 1 Autophagy is a cellular pathway involved in protein and organelle degradation, which is likely to represent an innate adaptation to starvation. In times of nutrient deficiency, the cell can self-digest and recycle some nonessential components through nonselective autophagy, thus sustaining minimal growth requirements until a food source becomes available. Over recent years, autophagy has been implicated in an increasing number of clinical scenarios, notably infectious diseases, cancer, neurodegenerative diseases, and autoimmunity. The recent identification of the importance of autophagy genes in the genetic susceptibility to Crohn ’ s disease suggests that a selective autophagic response may play a crucial role in the pathogenesis of common complex immune-mediated diseases. In this review, we discuss the autophagic mechanisms, their molecular regulation, and summarize their clinical relevance. This progress has led to great interest in the therapeutic potential of manipulation of both selective and nonselective autophagy in established disease. INTRODUCTION The ability to adapt to environmental change is essential for sur- Autophagy encompasses several distinct processes involving vival. This is true for the organism as a whole and for individual the delivery of portions of the cytoplasm to the lysosome for cells alike. -

Katalog 2015 Cover Paul Lin *Hinweis Förderung.Indd
Product List 2015 WE LIVE SERVICE Certificates quartett owns two productions sites that are certified according to EN ISO 9001:2008 Quality management systems - Requirements EN ISO 13485:2012 + AC:2012 Medical devices - Quality management systems - Requirements for regulatory purposes GMP Conformity Our quality management guarantees products of highest quality! 2 Foreword to the quartett product list 2015 quartett Immunodiagnostika, Biotechnologie + Kosmetik Vertriebs GmbH welcomes you as one of our new business partners as well as all of our previous loyal clients. You are now member of quartett´s worldwide customers. First of all we would like to introduce ourselves to you. Founded as a family-run company in 1986, quartett ensures for more than a quarter of a century consistent quality of products. Service and support of our valued customers are our daily businesses. And we will continue! In the end 80´s quartett offered radioimmunoassay and enzyme immunoassay kits from different manufacturers in the USA. In the beginning 90´s the company changed its strategy from offering products for routine diagnostic to the increasing field of research and development. Setting up a production plant in 1997 and a second one in 2011 supported this decision. The company specialized its product profile in the field of manufacturing synthetic peptides for antibody production, peptides such as protease inhibitors, biochemical reagents and products for histology, cytology and immunohistology. All products are exclusively manufactured in Germany without outsourcing any production step. Nowadays, we expand into all other diagnostic and research fields and supply our customers in universities, government institutes, pharmaceutical and biotechnological companies, hospitals, and private doctor offices. -

Characterization of Gf a Drosophila Trimeric G Protein Alpha Subunit
Characterization of Gf a Drosophila trimeric G protein alpha subunit Naureen Quibria Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2012 © 2012 Naureen Quibria All rights reserved Abstract Characterization of Gf a Drosophila trimeric G-protein alpha subunit Naureen Quibria In the morphogenesis of tissue development, how coordination of patterning and growth achieve the correct organ size and shape is a principal question in biology. Efficient orchestrating mechanisms are required to achieve this and cells have developed sophisticated systems for reception and interpretation of the multitude of extracellular stimuli to which they are exposed. Plasma membrane receptors play a key role in the transmission of such signals. G-protein coupled receptors (GPCRs) are the largest class of cell surface receptors that respond to an enormous diversity of extracellular stimuli, and are critical mediators of cellular signal transduction in eukaryotic organisms. Signaling through GPCRs has been well characterized in many biological contexts. While they are a major class of signal transducers, there are not many defined instances where GPCRs have been implicated in the process of development to date. The Drosophila wing provides an ideal model system to elucidate and address the role of GPCRs in development, as its growth is regulated by a small number of conserved signaling pathways. In my thesis work, I address the role of a trimeric G alpha protein in Drosophila, Gαf, and what part it may play in development. In particular, I explore the role of Gαf as an alpha subunit of a trimeric complex, to determine what heptahelical receptors might act as its cognate receptor. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Role of Excess Inorganic Pyrophosphate in Primer-Extension Genotyping Assays
Methods Role of Excess Inorganic Pyrophosphate in Primer-Extension Genotyping Assays Ming Xiao,1 Angie Phong,1 Kristen L. Lum,1 Richard A. Greene,2 Philip R. Buzby,2,3 and Pui-Yan Kwok1,4,5 1Cardiovascular Research Institute, University of California, San Francisco, San Francisco, California 94143-0130, USA; 2PerkinElmer Life and Analytical Sciences, Inc., Boston, Massachusetts 02118-2512, USA We have developed and genotyped >15,000 SNP assays by using a primer extension genotyping assay with fluorescence polarization (FP) detection. Although the 80% success rate of this assay is similar to those of other SNP genotyping assays, we wanted to determine the reasons for the failures and find ways to improve the assay. We observed that the failed assays fell into three general patterns: PCR failure, excess of heterozygous genotypes, and loss of FP signal for one of the dye labels. After analyzing several hundred failed assays, we concluded that 5% of the assays had PCR failure and had to be redesigned. We also discovered that the other two categories of failures were due to misincorporation of one of the dye-terminators during the primer extension reaction as a result of primer shortening with a reverse reaction involving inorganic pyrophosphate, and to the quenching of R110-terminator after its incorporation onto the SNP primer. The relatively slow incorporation of R110 acycloterminators by AcycloPol compounds the problem with the R110 label. In this report, we describe the source of the problems and simple ways to correct these problems by adding pyrophosphatase, using quenching as part of the analysis, and replacing R110 by Texas red as one of the dye labels. -

Neuropeptidergic Signaling Partitions Arousal Behaviors in Zebrafish
3142 • The Journal of Neuroscience, February 26, 2014 • 34(9):3142–3160 Behavioral/Cognitive Neuropeptidergic Signaling Partitions Arousal Behaviors in Zebrafish Ian G. Woods,1,2 David Schoppik,2 Veronica J. Shi,2 Steven Zimmerman,2 Haley A. Coleman,1 Joel Greenwood,3 Edward R. Soucy,3 and Alexander F. Schier2,3 1Department of Biology, Ithaca College, Ithaca, New York 14850, and 2Department of Molecular and Cellular Biology and 3Center for Brain Science, Harvard University, Cambridge, Massachusetts 02138 Animals modulate their arousal state to ensure that their sensory responsiveness and locomotor activity match environmental demands. Neuropeptides can regulate arousal, but studies of their roles in vertebrates have been constrained by the vast array of neuropeptides and their pleiotropic effects. To overcome these limitations, we systematically dissected the neuropeptidergic modulation of arousal in larval zebrafish. We quantified spontaneous locomotor activity and responsiveness to sensory stimuli after genetically induced expression of seven evolutionarily conserved neuropeptides, including adenylate cyclase activating polypeptide 1b (adcyap1b), cocaine-related and amphetamine-related transcript (cart), cholecystokinin (cck), calcitonin gene-related peptide (cgrp), galanin, hypocretin, and nocicep- tin. Our study reveals that arousal behaviors are dissociable: neuropeptide expression uncoupled spontaneous activity from sensory responsiveness, and uncovered modality-specific effects upon sensory responsiveness. Principal components analysis and phenotypic clustering revealed both shared and divergent features of neuropeptidergic functions: hypocretin and cgrp stimulated spontaneous locomotor activity, whereas galanin and nociceptin attenuated these behaviors. In contrast, cart and adcyap1b enhanced sensory respon- siveness yet had minimal impacts on spontaneous activity, and cck expression induced the opposite effects. Furthermore, hypocretin and nociceptin induced modality-specific differences in responsiveness to changes in illumination. -

DDX5 Targets Tissue-Specific Rnas to Promote Intestine Tumorigenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.25.006668; this version posted March 26, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. DDX5 targets tissue-specific RNAs to promote intestine tumorigenesis Nazia Abbasi1,4, Tianyun Long1,4, Yuxin Li1, Evelyn Ma1, Brian A. Yee1, Parth R. Patel1, Ibrahim M SayeD2, Nissi Varki2, Soumita Das2, PraDipta Ghosh1, 3, Gene W. Yeo1, WenDy J.M. Huang1,5 1 Department of Cellular anD Molecular MeDicine, University of California San Diego, La Jolla, CA 2 Department of Pathology, University of California San Diego, La Jolla, CA 3 Department of MeDicine, University of California San Diego, La Jolla, CA 4 These authors contributeD equally 5 CorresponDing author email: [email protected] Abstract Tumorigenesis in Different segments of the intestinal tract involves tissue-specific oncogenic Drivers. In the colon, complement component 3 (C3) activation is a major contributor to inflammation anD malignancies. By contrast, tumorigenesis in the small intestine involves fatty aciD-binding protein 1 (FABP1). However, little is known of the upstream mechanisms Driving their expressions in Different segments of the intestinal tract. Here, we report that an RNA binDing protein DDX5 augments C3 and FABP1 expressions post-transcriptionally to promote tumorigenesis in the colon anD small intestine, respectively. Mice with epithelial-specific knockout of DDX5 are protecteD from intestine tumorigenesis. The iDentification of DDX5 as the common upstream regulator of tissue-specific oncogenic molecules proviDes a new therapeutic target for intestine cancers. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -
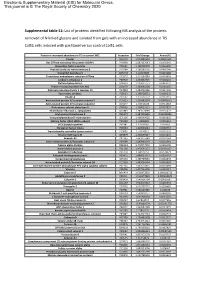
Supplemental Table S1
Electronic Supplementary Material (ESI) for Molecular Omics. This journal is © The Royal Society of Chemistry 2020 Supplemental table S1: List of proteins identified following MS analysis of the proteins removed of N-linked glycans and isolated from gels with an increased abundance in TIS Cal51 cells induced with paclitaxel versus control Cal51 cells. Protein in increased abundance in TIS vs control WCL Accession Fold Change Anova (P) Plectin Q15149 1.073855593 0.00691631 Ras GTPase-activating-like protein IQGAP1 P46940 1.087337643 0.0176342 Elongation factor1-gamma P26641 1.138709703 0.0116496 Peptidyl-prolyl cis-transisomerase B P23284 1.188383105 0.0436246 Dipeptidyl peptidase 3 Q9NY33 1.20163605 0.0215448 Transitional endoplasmic reticulum ATPase P55072 1.214194884 0.0449691 Carbonic anhydrase 2 P00918 1.232852325 0.0158141 Clathrin heavy chain 1 Q00610 1.239621773 0.0463237 Protein transport protein Sec 31A O94979 1.263565104 0.0284155 Aldo-ketoreductase family 1 member C1 Q04828 1.282092186 0.0324406 Spermidine synthase P19623 1.298728621 0.0196232 Plastin-3 P13797 1.310756772 0.0161319 Actin-related protein 2/3 complex subunit 5 O15511 1.333483524 0.00476923 Actin-related protein 2/3 complex subunit 2 O15144 1.35416168 0.0411018 Proteasome subunit alpha type-5 P28066 1.358015551 0.0337657 Thioredoxin reductase 1, cytoplasmic Q16881 1.383670089 0.0235472 Acyl-protein thioesterase 2 O95372 1.387415589 0.00233899 Isoaspartylpeptidase/L-asparaginase Q7L266 1.408149002 0.0319602 Splicing factor U2AF 65kDa subunit P26368 1.41489991 0.0256619 -

Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/Mtor Cascade Inhibitors: How Mutations Can Result in Therapy Resistance and How to Overcome Resistance
www.impactjournals.com/oncotarget/ Oncotarget, October, Vol.3, No 10 Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR Cascade Inhibitors: How Mutations Can Result in Therapy Resistance and How to Overcome Resistance James A. McCubrey1, Linda S. Steelman1, William H. Chappell1, Stephen L. Abrams1, Richard A. Franklin1, Giuseppe Montalto2, Melchiorre Cervello3, Massimo Libra4, Saverio Candido4, Grazia Malaponte4, Maria C. Mazzarino4, Paolo Fagone4, Ferdinando Nicoletti4, Jörg Bäsecke5, Sanja Mijatovic6, Danijela Maksimovic- Ivanic6, Michele Milella7, Agostino Tafuri8, Francesca Chiarini9, Camilla Evangelisti9, Lucio Cocco10, Alberto M. Martelli9,10 1 Department of Microbiology and Immunology, Brody School of Medicine at East Carolina University, Greenville, NC, USA 2 Department of Internal Medicine and Specialties, University of Palermo, Palermo, Italy 3 Consiglio Nazionale delle Ricerche, Istituto di Biomedicina e Immunologia Molecolare “Alberto Monroy”, Palermo, Italy 4 Department of Bio-Medical Sciences, University of Catania, Catania, Italy 5 Department of Medicine, University of Göttingen, Göttingen, Germany 6 Department of Immunology, Instititue for Biological Research “Sinisa Stankovic”, University of Belgrade, Belgrade, Serbia 7 Regina Elena National Cancer Institute, Rome, Italy 8 Sapienza, University of Rome, Department of Cellular Biotechnology and Hematology, Rome, Italy 9 Institute of Molecular Genetics, National Research Council-Rizzoli Orthopedic Institute, Bologna, Italy. 10 Department of Biomedical and Neuromotor Sciences, University of Bologna, Bologna, Italy Correspondence to: James A. McCubrey, email: [email protected] Keywords: Targeted Therapy, Therapy Resistance, Cancer Stem Cells, Raf, Akt, PI3K, mTOR Received: September 12, 2012, Accepted: October 18, 2012, Published: October 20, 2012 Copyright: © McCubrey et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog.