(MST) Analyses of Three Connecticut Watersheds of Long Island Sound
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
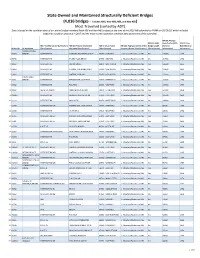
State Owned and Maintained Structurally Deficient Bridges
State Owned and Maintained Structurally Deficient Bridges (4,016 bridges - includes NHS, Non-NHS, NBI, and Non-NBI) Most Travelled (sorted by ADT) Data is based on the condition data of our entire bridge inventory (both NBI and Non-NBI bridges) at the time of the 2018 NBI submittal to FHWA on 03/15/18, which included inspection condition data due in 2017 and the most recent inspection condition data at the time of the submittal NBI 29: Average NBI 112: NBIS Daily Traffic (ADT) NBI 27: Year NBI 7: Facility Carried by Structure NBI 6A: Feature Intersected: NBI 4: City or Town NBI 104: Highway System of the Bridge Length [General Built [General Bridge No. Bridge Name [Classification] Narrative [Classification] [Classification] Inventory Route [Classification] [Classification] Information] Information] YANKEE DOODLE 1 00059 BRIDGE INTERSTATE-95 NORWALK RV,HENDRICKS AVE 56060 - NORWALK 1 - Structure/Route is on NHS Yes 145000 1957 2 06798 INTERSTATE-95 STUBBY PLAIN BROOK 47535 - MILFORD 1 - Structure/Route is on NHS No 137500 1958 3 00162 INTERSTATE-95 METRO NORTH 82870 - WEST HAVEN 1 - Structure/Route is on NHS Yes 136400 1956 4 03093 INTERSTATE 91 N FRONT ST & QUINN RIVER 52070 - NEW HAVEN 1 - Structure/Route is on NHS Yes 133900 1964 5 03094 INTERSTATE-91 AMTRAK RAILROAD 52070 - NEW HAVEN 1 - Structure/Route is on NHS Yes 133900 1965 BYRAM RIVER 6 00001 BRIDGE INTERSTATE-95 BYRAM RIVER,S WATER ST 33620 - GREENWICH 1 - Structure/Route is on NHS Yes 131600 1958 7 00062 INTERSTATE-95 ROUTE 33 83500 - WESTPORT 1 - Structure/Route is on NHS -

Department of Environmental Protection Inland Waters And
Regulations of Connecticut State Agencies TITLE 26. Fisheries & Game Agency Department of Environmental Protection Subject Inland Waters and Marine District Defined Section § 26-108-1 CONTENTS Sec. 26-108-1. Inland waters and marine district defined Revised: 2015-3-6 R.C.S.A. § 26-108-1 - I- Regulations of Connecticut State Agencies TITLE 26. Fisheries & Game Department of Environmental Protection §26-108-1 Inland Waters and Marine District Defined Sec. 26-108-1. Inland waters and marine district defined The following lines across streams flowing into Long Island Sound, Fisher’s Island Sound, Little Narragansett Bay and tributaries thereof shall be known as the inland-marine demarcation lines above which lines such water shall be known as the “inland district” and below which lines such water shall be known as the “marine district”: FAIRFIELD COUNTY Byram River, Greenwich marine district—up to and including the railroad crossing inland district—all waters above Horse Neck Brook, Greenwich marine district—none inland district—above junction with mouth of harbor Indian Harbor, Greenwich marine district—up to and including the first railroad crossing inland district—all waters above Mianus River, Greenwich marine district—below dam just above Boston Post Road inland district—all waters above Rippowam River, Stamford marine district—up to and including the first railroad crossing inland district—all waters above Noroton River, Stamford-Darien marine district—up to and including Boston Post Road inland district—all waters above Goodwives River, -

CT DEEP 2018 FISHING REPORT NUMBER 1 Channel Catfish (Ictalurus Punctatus) 4/26/2018 Brown Trout (Salmo Trutta)
CT DEEP 2018 FISHING REPORT NUMBER 1 Channel catfish (Ictalurus punctatus) 4/26/2018 Brown Trout (Salmo trutta) YOU CAN FIND US DIRECTLY ON FACEBOOK. This page features a variety of information on fishing, hunting, and wildlife watching in Connecticut. The address is www.facebook.com/CTFishandWildlife. INLAND REPORT OPENING DAY – We had a short blast of warm air temperatures that gave anglers a comfortable Opening Day, however, water temperatures were very cold, possibly contributing to difficult catching for many. Fisheries staff were out at eight of the twelve Trout Parks were stocked on Opening Day and the many kids Connecticut’s Trout & Salmon Stamp: Connecticut present enjoyed helping us stock. Catch percentage has implemented a Trout and Salmon Stamp. 100% was from 60 to 80% at a number of the Trout Parks of the revenue from your investment comes to the including Stratton Brook, Black Rock, Kent Falls, DEEP Bureau of Natural Resources for Fisheries Chatfield Hollow, Valley Falls Park, Southford Falls, and programs. Great Hollow. Many other locations, both river and The Trout and Salmon Stamp is $5 for anyone age 18 stream as well as lake and pond did not give up their or older, including those 65 or older, and $3 for CT recently stocked trout so easily. residents age 16-17. The Stamp is required for the harvest (keeping) of Over 300,000 trout were stocked before Opening Day trout or salmon. into nearly 100 lakes and ponds and over 120 rivers The Stamp is required to FISH in one of these places: and streams located throughout Connecticut. -

LISS 3.3.Qxd
RestoringRestoring LongLong CONNECTICUT Connecticut Quinnipiac River River IslandIsland Thames Sound’s River Sound’s Housatonic River Stonington HabitatsHabitats Old Saybrook COMPLETED RESTORATION SITES IN PROGRESS RESTORATION SITES POTENTIAL RESTORATION SITES PROJECT BOUNDARY RIVER LONG ISLAND SOUND Greenwich 2002 RESTORATION SITES Southold BLUE INDICATES COMPLETED SITE – CONSTRUCTION ON THE PROJECT IS FINISHED, BUT MONITORING MAY BE ON-GOING GREEN INDICATES IN PROGRESS SITE– SOME PHASE OF THE PROJECT IS UNDERWAY, E.G. APPLYING FOR FUNDING, DESIGN, OR CONSTRUCTION BLACK INDICATES POTENTIAL SITE – A RESTORATION PROJECT HAS BEEN IDENTIFIED, NO ACTION TAKEN YET MOUNT VERNON RYE BOLDFACE IN ALL COLORS INDICATES HIGH-RANKED SITES Rye Glover Field (FW) Beaver Swamp Brook (FW) Beaver Swamp Brook/Cowperwood site (FW) Brookhaven NEW ROCHELLE Blind Brook (FW) Echo Bay (TW/SR/IF/RI) Edith G. Read Wildlife Sanctuary (TW/F/EE/FW) CONNECTICUT Former Dickerman’s Pond (FW) Marshlands Conservancy (TW/F/IF) Farm River (TW) EW ORK Nature Study Woods (F/FW) Farm River tributary/Edgemere Rd. (TW) N Y Pryer Manor Marsh (TW) SMITHTOWN BRANFORD Morris Creek/Sibley Lane (TW) Callahan’s Beach (CB) Branford River STP (TW) New Haven Airport (TW) Bronx BRONX NORTH HEMPSTEAD Fresh Pond (FW/F/BD) Branford R./Christopher Rd. (TW) Nissequogue Bronx Oyster Reefs (SR) Baxter Estates Pond (FW) Harrison Pond Town Park (FW/RMC/TW/F) Branford R./St. Agnes Cemetery (TW) EAST LYME NEW YORK Bronx River mouth (TW/F/RMC) Hempstead Harbor (EE/IF/TW) Landing Avenue Town Park (TW) Branford R./Hickory Rd. (TW) Brides Brook Culvert (RMC/TW) River Bronx River Trailway (TW/FW/F/RMC) Lake Success (FW) Long Beach (BD) Branford R. -

Connecticut Watersheds
Percent Impervious Surface Summaries for Watersheds CONNECTICUT WATERSHEDS Name Number Acres 1985 %IS 1990 %IS 1995 %IS 2002 %IS ABBEY BROOK 4204 4,927.62 2.32 2.64 2.76 3.02 ALLYN BROOK 4605 3,506.46 2.99 3.30 3.50 3.96 ANDRUS BROOK 6003 1,373.02 1.03 1.04 1.05 1.09 ANGUILLA BROOK 2101 7,891.33 3.13 3.50 3.78 4.29 ASH CREEK 7106 9,813.00 34.15 35.49 36.34 37.47 ASHAWAY RIVER 1003 3,283.88 3.89 4.17 4.41 4.96 ASPETUCK RIVER 7202 14,754.18 2.97 3.17 3.31 3.61 BALL POND BROOK 6402 4,850.50 3.98 4.67 4.87 5.10 BANTAM RIVER 6705 25,732.28 2.22 2.40 2.46 2.55 BARTLETT BROOK 3902 5,956.12 1.31 1.41 1.45 1.49 BASS BROOK 4401 6,659.35 19.10 20.97 21.72 22.77 BEACON HILL BROOK 6918 6,537.60 4.24 5.18 5.46 6.14 BEAVER BROOK 3802 5,008.24 1.13 1.22 1.24 1.27 BEAVER BROOK 3804 7,252.67 2.18 2.38 2.52 2.67 BEAVER BROOK 4803 5,343.77 0.88 0.93 0.94 0.95 BEAVER POND BROOK 6913 3,572.59 16.11 19.23 20.76 21.79 BELCHER BROOK 4601 5,305.22 6.74 8.05 8.39 9.36 BIGELOW BROOK 3203 18,734.99 1.40 1.46 1.51 1.54 BILLINGS BROOK 3605 3,790.12 1.33 1.48 1.51 1.56 BLACK HALL RIVER 4021 3,532.28 3.47 3.82 4.04 4.26 BLACKBERRY RIVER 6100 17,341.03 2.51 2.73 2.83 3.00 BLACKLEDGE RIVER 4707 16,680.11 2.82 3.02 3.16 3.34 BLACKWELL BROOK 3711 18,011.26 1.53 1.65 1.70 1.77 BLADENS RIVER 6919 6,874.43 4.70 5.57 5.79 6.32 BOG HOLLOW BROOK 6014 4,189.36 0.46 0.49 0.50 0.51 BOGGS POND BROOK 6602 4,184.91 7.22 7.78 8.41 8.89 BOOTH HILL BROOK 7104 3,257.81 8.54 9.36 10.02 10.55 BRANCH BROOK 6910 14,494.87 2.05 2.34 2.39 2.48 BRANFORD RIVER 5111 15,586.31 8.03 8.94 9.33 9.74 -

The Fishway at the East Haven Dam on the Farm River (First Upstream
Easygrants ID: 39391 National Fish and Wildlife Foundation NFWF Project ID: 1401.13.039391 LI Sound Futures Fund 2013 - Planning All Types and Water Quality Monitoring - Submit Final Programmatic Report (New Metrics) Grantee Organization: Hammonasset Chapter #446, Trout Unlimited, Inc. Project Title: Pages Millpond Fishway Design, Farm River (CT) Project Period 10/01/2013 - 6/30/2015 Project Location Pages Millpond Dam, North Branford and East Haven Dam, East Haven, Farm River, CT. Description (from Proposal) Project Finalize a plan for two fishways restoring free fish passage to 5.35 river miles to increase spawning Summary (from habitat for alewife, blueback herring, American eel, and sea run brown trout. Proposal) Summary of The fishway at the East Haven Dam on the Farm River (first upstream impoundment from Accomplishments Long Island Sound) was designed and constructed by the South Central Connecticut Regional Water Authority. This fishway allows access by diadromous fish, at essentially all water levels, to 1.75 river miles of the Farm River up to the Pages Millpond Dam. The final design and permitting of the Pages Millpond Dam fishway was completed. A determination that the project was eligible under Category 1 of the Connecticut General Permit was issued by the U.S. Army Corps of Engineers on April 17, 2014. This determination is valid until the General Permit expires on 07-15-2016. It is anticipated that this determination can be easily renewed when a new General Permit is issued. A Dam Safety construction permit was issued on October 22, 2014 by the CT DEEP authorizing the project pursuant to Section 22a-403 of the CGS. -

CONNECTICUT Estbrook Harbor
280 ¢ U.S. Coast Pilot 2, Chapter 8 Chapter 2, Pilot Coast U.S. 72°30'W 72°W Chart Coverage in Coast Pilot 2—Chapter 8 Hartford NOAA’s Online Interactive Chart Catalog has complete chart coverage http://www.charts.noaa.gov/InteractiveCatalog/nrnc.shtml 12378 73°W CONNECTICUT Norwich 12372 41°30'N C O 12377 THAMES RIVER N N E C T I C U T R I V E R 12375 New London 12372 12354 Essex HOUSATONIC RIVER New Haven NIANTIC BAY 13213 12371 12373 12374 Westbrook Harbor 13211 Branford Harbor Guilford Harbor 12372 BLOCK ISLAND SOUND 12358 Orient Point 12370 LONG ISLAND SOUND 41°N 12362 Port Je erson L ONG ISLAND NORTH ATLANTIC OCEAN 19 SEP2021 19 SEP 2021 U.S. Coast Pilot 2, Chapter 8 ¢ 281 Eastern Long Island Sound (1) This chapter describes the eastern portion of Long by small vessels when meeting unfavorable weather or Island Sound following the north shore from Thames reaching the eastern part of the sound. Small vessels can River to and including the Housatonic River and then select anchorage eastward or westward of Kelsey Point the south shore from Orient Point to and including Port Breakwater, also in Duck Island Roads. Off Madison Jefferson. Also described are the Connecticut River; the there is anchorage sheltered from northerly winds. New ports of New London, New Haven and Northville; and the Haven Harbor is an important harbor of refuge. more important fishing and yachting centers on Niantic (11) Several general anchorages are in Long Island River and Bay, Westbrook Harbor, Guilford Harbor, Sound. -

Schenob Brook
Sages Ravine Brook Schenob BrookSchenob Brook Housatonic River Valley Brook Moore Brook Connecticut River North Canaan Watchaug Brook Scantic RiverScantic River Whiting River Doolittle Lake Brook Muddy Brook Quinebaug River Blackberry River Hartland East Branch Salmon Brook Somers Union Colebrook East Branch Salmon Brook Lebanon Brook Fivemile RiverRocky Brook Blackberry RiverBlackberry River English Neighborhood Brook Sandy BrookSandy Brook Muddy Brook Freshwater Brook Ellis Brook Spruce Swamp Creek Connecticut River Furnace Brook Freshwater Brook Furnace Brook Suffield Scantic RiverScantic River Roaring Brook Bigelow Brook Salisbury Housatonic River Scantic River Gulf Stream Bigelow Brook Norfolk East Branch Farmington RiverWest Branch Salmon Brook Enfield Stafford Muddy BrookMuddy Brook Factory Brook Hollenbeck River Abbey Brook Roaring Brook Woodstock Wangum Lake Brook Still River Granby Edson BrookEdson Brook Thompson Factory Brook Still River Stony Brook Stony Brook Stony Brook Crystal Lake Brook Wangum Lake Brook Middle RiverMiddle River Sucker BrookSalmon Creek Abbey Brook Salmon Creek Mad RiverMad River East Granby French RiverFrench River Hall Meadow Brook Willimantic River Barkhamsted Connecticut River Fenton River Mill Brook Salmon Creek West Branch Salmon Brook Connecticut River Still River Salmon BrookSalmon Brook Thompson Brook Still River Canaan Brown Brook Winchester Broad BrookBroad Brook Bigelow Brook Bungee Brook Little RiverLittle River Fivemile River West Branch Farmington River Windsor Locks Willimantic River First -

Connecticut Sea Grant Project Report
1 CONNECTICUT SEA GRANT PROJECT REPORT Please complete this progress or final report form and return by the date indicated in the emailed progress report request from the Connecticut Sea Grant College Program. Fill in the requested information using your word processor (i.e., Microsoft Word), and e-mail the completed form to Dr. Syma Ebbin [email protected], Research Coordinator, Connecticut Sea Grant College Program. Do NOT mail or fax hard copies. Please try to address the specific sections below. If applicable, you can attach files of electronic publications when you return the form. If you have questions, please call Syma Ebbin at (860) 405-9278. Please fill out all of the following that apply to your specific research or development project. Pay particular attention to goals, accomplishments, benefits, impacts and publications, where applicable. Project #: __R/CE-34-CTNY__ Check one: [ ] Progress Report [ x ] Final report Duration (dates) of entire project, including extensions: From [March 1, 2013] to [August 28, 2015 ]. Project Title or Topic: Comparative analysis and model development for determining the susceptibility to eutrophication of Long Island Sound embayment. Principal Investigator(s) and Affiliation(s): 1. Jamie Vaudrey, Department of Marine Sciences, University of Connecticut 2. Charles Yarish, Department of Ecology & Evolutionary Biology, Department of Marine Sciences, University of Connecticut 3. Jang Kyun Kim, Department of Marine Sciences, University of Connecticut 4. Christopher Pickerell, Marine Program, Cornell Cooperative Extension of Suffolk County 5. Lorne Brousseau, Marine Program, Cornell Cooperative Extension of Suffolk County A. COLLABORATORS AND PARTNERS: (List any additional organizations or partners involved in the project.) Justin Eddings, Marine Program, Cornell Cooperative Extension of Suffolk County Michael Sautkulis, Cornell Cooperative Extension of Suffolk County Veronica Ortiz, University of Connecticut Jeniam Foundation (Tripp Killin, Exec. -

The Connecticut River May Produce Seed Oysters Once Again by Timothy C
The Connecticut River May Produce Seed Oysters Once Again By Timothy C. Visel Over a century ago, Connecticut Prevents Self Purification of My graduate research at the oystermen were developing an Water Courses, delivered to a University of Rhode Island industry that would become world convention of the American involves developing the shellfish renowned for its ability to produce Health Association in 1922, Dr. management program for Old large quantities of seed oysters. Thurlow Nelson examined this Saybrook, Connecticut. Old They observed that the rivers and decline, nothing the outpouring of Saybrook is situated at the mouth streams that flow into Long Island industrial waste incident to the of the Connecticut River and has Sound created a brackish element war. “Let present conditions several miles of shoreline that was especially suitable for continue,” he said, “and in ten bordering Long Island Sound to oyster growing. years the oyster will only be a the south and the Connecticut memory.” River to the west. In the last two The technique they used was years, several shellfish surveys called “natural growth.” It was Forty-five years after Dr. Nelson’s have revealed two large natural based upon the realization that address, production of oysters oyster beds in the lower young oysters, called “spat,” from Long Island Sound had Connecticut River. attached themselves in great fallen to only 40,000 bushels, or numbers on oyster shells less than 3 percent of earlier In the summer lf 1082, Old discarded over oyster beds. harvests. Saybrook transplanted Capitalizing on the oyster’s approximately 100 bushes of spawning characteristics and the In 1967, Connecticut’s oyster oysters from polluted areas in the need of a clean surface upon industry received a government Connecticut River to offshore which that spat must be affixed, sponsored resource disaster grant areas. -

Ct Deep 2018 Spring Stocking Trout
CT DEEP 2018 SPRING STOCKING TROUT YOU CAN FIND US DIRECTLY ON FACEBOOK. This page features a variety of information on fishing, hunting, and wildlife watching in Connecticut. The address is www.facebook.com/CTFishandWildlife. DEEP’s 2018 spring trout stockings were completed on May 25. Prior to Opening Day, approximately 306,000 trout were stocked into nearly 100 lakes and ponds and over 120 rivers and streams located throughout Connecticut. DEEP’s in-season stockings are now underway with nearly 240,000 additional trout scheduled to be stocked between Opening Day and the middle of May. On July 3 DEEP conducted its annual trout stocking on the Farmington River and fall stockings will begin in late August or early September if weather and flows cooperate. The following area was stocked on July 3 (updated 7/5/2018) RIVERS AND STREAMS: Just in time for the Fourth of July holiday, The Farmington River TMA (the West Branch Farmington River from the Goodwin Dam down to the upper boundary of the year-round catch-and-release area) was stocked with 1,800 Brown Trout (12 inch and up fish). The following areas were stocked the week of 5/21 - 5/25 (updated 5/25/2018) RIVERS AND STREAMS: In eastern Connecticut; Fenton River, Scantic River (Upper and lower), Natchaug River, Quinebaug River, Farm River (Lower), Jeremy River, Coginchaug River, the Salmon River & Salmon River TMA, Shetucket River and the Moosup River & the Moosup River TMA. In western Connecticut; Pequabuck River, Pequonnock River (Trumbull Basin, Open and Beardsley Park), the lower Naugatuck River (Waterbury – Beacon Falls), Weekeepeemee River, Little River (Oxford), Hop Brook, Nonewaug River and Pomperaug River. -

Long Island Sound Habitat Restoration Initiative
LONG ISLAND SOUND HABITAT RESTORATION INITIATIVE ANNUAL SUMMARY FOR THE YEAR 2000 Technical Support for Coastal Habitat Restoration Prepared by: Office of Long Island Sound Programs 79 Elm Street Hartford, CT 06106 860-424-3034 http://dep.state.ct.us and New York State Department of Environmental Conservation 205 N. Belle Meade Road East Setauket, NY 11733 631-444-0430 www.dec.state.ny.us ANNUAL SUMMARY FOR THE YEAR 2000 TABLE OF CONTENTS LONG ISLAND SOUND HABITAT RESTORATION INITIATIVE -- ANNUAL SUMMARY FOR THE YEAR 2000 ............................................................... S-1 BACKGROUND..........................................................................................................................S-1 2000 PROGRESS REPORT ........................................................................................................S-2 TIDAL WETLAND RESTORATION ..........................................................................................S-3 Nott Island .............................................................................................................................. S-6 Hammonasset State Park, Madison ................................................................................... S-7 Farm River Tributary Marsh................................................................................................. S-8 Centre Island Wetland ......................................................................................................... S-9 Edith Read Natural Park and Wildlife Sanctuary...........................................................