Ungulata (Hoefdieren)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inclusion of Asiatic Mammal Species on Cms Appendices
Convention on the Conservation of Migratory Species of Wild Animals Secretariat provided by the United Nations Environment Programme 14 th MEETING OF THE CMS SCIENTIFIC COUNCIL Bonn, Germany, 14-17 March 2007 CMS/ScC14/Doc.13 Agenda item 6(a) DRAFT PROPOSALS FOR THE INCLUSION OF ASIATIC MAMMAL SPECIES ON CMS APPENDICES (Prepared by the Secretariat) 1. The four draft proposals for the amendment of CMS Appendices attached to this note have been prepared by the Institut Royal des Sciences Naturelles de Belgique and have been submitted by Dr. Pierre Devillers, Scientific Councillor for the European Community and vice-chairman of the Scientific Council. 2. Preparation of these draft proposals is undertaken within the Central Eurasian Aridland Concerted Action and associated Cooperative Action approved by the 8 th Meeting of the Conference of the Parties to CMS (Recommendation 8.23), covering threatened migratory large mammals of the temperate and cold deserts, semi-deserts, steppes and associated mountains of Central Asia, the Northern Indian sub-continent, Western Asia, the Caucasus and Eastern Europe. 3. In particular, Rec. 8.23 “encourages Range States and other interested Parties to prepare, in cooperation with the Scientific Council and the Secretariat, the necessary proposals to include in Appendix I or Appendix II threatened species that would benefit from the Action”. For reasons of economy, documents are printed in a limited number, and will not be distributed at the meeting. Delegates are kindly requested to bring their copy to the meeting and not to request additional copies. DRAFT PROPOSAL FOR INCLUSION OF SPECIES ON THE APPENDICES OF THE CONVENTION ON THE CONSERVATION OF MIGRATORY SPECIES OF WILD ANIMALS Proposal to add in Appendix I Pantholops hodgsonii Document largely based on the species information provided in IUCN Redlist of Threatened Species database (2006) February 2007 2 1. -

Boselaphus Tragocamelus</I>
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- Published Research US Geological Survey 2008 Boselaphus tragocamelus (Artiodactyla: Bovidae) David M. Leslie Jr. U.S. Geological Survey, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/usgsstaffpub Leslie, David M. Jr., "Boselaphus tragocamelus (Artiodactyla: Bovidae)" (2008). USGS Staff -- Published Research. 723. https://digitalcommons.unl.edu/usgsstaffpub/723 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- Published Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. MAMMALIAN SPECIES 813:1–16 Boselaphus tragocamelus (Artiodactyla: Bovidae) DAVID M. LESLIE,JR. United States Geological Survey, Oklahoma Cooperative Fish and Wildlife Research Unit and Department of Natural Resource Ecology and Management, Oklahoma State University, Stillwater, OK 74078-3051, USA; [email protected] Abstract: Boselaphus tragocamelus (Pallas, 1766) is a bovid commonly called the nilgai or blue bull and is Asia’s largest antelope. A sexually dimorphic ungulate of large stature and unique coloration, it is the only species in the genus Boselaphus. It is endemic to peninsular India and small parts of Pakistan and Nepal, has been extirpated from Bangladesh, and has been introduced in the United States (Texas), Mexico, South Africa, and Italy. It prefers open grassland and savannas and locally is a significant agricultural pest in India. It is not of special conservation concern and is well represented in zoos and private collections throughout the world. DOI: 10.1644/813.1. -

O18934 Plard, F., Bonenfant, C. and Gaillard, JM 2011
Oikos O18934 Plard, F., Bonenfant, C. and Gaillard, J. M. 2011. Re- visiting the allometry of antlers among deer species: male-male sexual competition as a driver. – Oikos 120: 601–606. Table A1. Data basis. Sexual size dimorphism (SSD) was measured by the residuals of the regression of male body mass against female body mass. Breeding group sizes (BGS) were divided into three categories: group of one or two animals (A), group of three, four and five animals (B), and groups larger than five animals (C). The different mating tactics (MT) were harem (H), territorial (T) and tending (F). Non-available mating tactics (NA) and monogamous species (M) are indicated. Sub-family Genus Species Common name Male Female SSD BGS MT Ref Antler Body Body length mass mass Cervinae Axis axis chital 845 89.5 39 0.58 C F 1,2,3 Cervinae Axis porcinus hog deer 399 41 31 0.07 C F 2,3,4,5 Cervinae Cervus albirostris white-lipped deer 1150 204 125 0.06 C H 1,6 Cervinae Cervus canadensis wapiti 1337 350 250 –0.21 C H 3,8,9 Cervinae Cervus duvaucelii barasingha 813 236 145 0.03 C H 1,3,10,11 Cervinae Cervus elaphus red deer 936 250 125 0.34 C H 1,2,3 Cervinae Cervus eldi Eld’s deer 971.5 105 67 0.12 C H 1,2,3 Cervinae Cervus nippon sika deer 480 52 37 0.1 B T 1,2,3,9,12,13 Cervinae Cervus timorensis Timor deer 675 95.5 53 0.29 C H 1,9 Cervinae Cervus unicolor sambar 1049 192 146 –0.18 B T 1,2,3,7 Cervinae Dama dama fallow deer 615 67 44 0.15 C T 1,2,3,14 Cervinae Elaphurus davidianus Père David’s deer 737 214 159 –0.17 C H 1,2,3 Muntiacinae Elaphodus cephalophus -

An Analysis of the Phylogenetic Relationship of Thai Cervids Inferred from Nucleotide Sequences of Protein Kinase C Iota (PRKCI) Intron
Kasetsart J. (Nat. Sci.) 43 : 709 - 719 (2009) An Analysis of the Phylogenetic Relationship of Thai Cervids Inferred from Nucleotide Sequences of Protein Kinase C Iota (PRKCI) Intron Kanita Ouithavon1, Naris Bhumpakphan2, Jessada Denduangboripant3, Boripat Siriaroonrat4 and Savitr Trakulnaleamsai5* ABSTRACT The phylogenetic relationship of five Thai cervid species (n=21) and four spotted deer (Axis axis, n=4), was determined based on nucleotide sequences of the intron region of the protein kinase C iota (PRKCI) gene. Blood samples were collected from seven captive breeding centers in Thailand from which whole genomic DNA was extracted. Intron1 sequences of the PRKCI nuclear gene were amplified by a polymerase chain reaction, using L748 and U26 primers. Approximately 552 base pairs of all amplified fragments were analyzed using the neighbor-joining distance matrix method, and 19 parsimony- informative sites were analyzed using the maximum parsimony approach. Phylogenetic analyses using the subfamily Muntiacinae as outgroups for tree rooting indicated similar topologies for both phylogenetic trees, clearly showing a separation of three distinct genera of Thai cervids: Muntiacus, Cervus, and Axis. The study also found that a phylogenetic relationship within the genus Axis would be monophyletic if both spotted deer and hog deer were included. Hog deer have been conventionally classified in the genus Cervus (Cervus porcinus), but this finding supports a recommendation to reclassify hog deer in the genus Axis. Key words: Thailand, the family Cervidae, protein kinase C iota (PRKCI) intron, phylogenetic tree, taxonomy INTRODUCTION 1987; Gentry, 1994). Cervids, or what is commonly called “deer,” are mostly characterized The family Cervidae is one of the most by antlers with a bony inner core and velvet skin specious families of artiodactyls, with an extensive cover. -

Fitzhenry Yields 2016.Pdf
Stellenbosch University https://scholar.sun.ac.za ii DECLARATION By submitting this dissertation electronically, I declare that the entirety of the work contained therein is my own, original work, that I am the sole author thereof (save to the extent explicitly otherwise stated), that reproduction and publication thereof by Stellenbosch University will not infringe any third party rights and that I have not previously in its entirety or in part submitted it for obtaining any qualification. Date: March 2016 Copyright © 2016 Stellenbosch University All rights reserved Stellenbosch University https://scholar.sun.ac.za iii GENERAL ABSTRACT Fallow deer (Dama dama), although not native to South Africa, are abundant in the country and could contribute to domestic food security and economic stability. Nonetheless, this wild ungulate remains overlooked as a protein source and no information exists on their production potential and meat quality in South Africa. The aim of this study was thus to determine the carcass characteristics, meat- and offal-yields, and the physical- and chemical-meat quality attributes of wild fallow deer harvested in South Africa. Gender was considered as a main effect when determining carcass characteristics and yields, while both gender and muscle were considered as main effects in the determination of physical and chemical meat quality attributes. Live weights, warm carcass weights and cold carcass weights were higher (p < 0.05) in male fallow deer (47.4 kg, 29.6 kg, 29.2 kg, respectively) compared with females (41.9 kg, 25.2 kg, 24.7 kg, respectively), as well as in pregnant females (47.5 kg, 28.7 kg, 28.2 kg, respectively) compared with non- pregnant females (32.5 kg, 19.7 kg, 19.3 kg, respectively). -

Review of Asian Species/Country Combinations Subject to Long-Standing Import Suspensions
Review of Asian species/country combinations subject to long-standing import suspensions (Version edited for public release) SRG 54 Prepared for the European Commission Directorate General Environment ENV.E.2. – Environmental Agreements and Trade by the United Nations Environment Programme World Conservation Monitoring Centre November, 2010 UNEP World Conservation Monitoring PREPARED FOR Centre 219 Huntingdon Road The European Commission, Brussels, Belgium Cambridge CB3 0DL DISCLAIMER United Kingdom Tel: +44 (0) 1223 277314 The contents of this report do not necessarily Fax: +44 (0) 1223 277136 reflect the views or policies of UNEP or Email: [email protected] Website: www.unep-wcmc.org contributory organisations. The designations employed and the presentations do not imply ABOUT UNEP-WORLD CONSERVATION the expressions of any opinion whatsoever on MONITORING CENTRE the part of UNEP, the European Commission or contributory organisations concerning the The UNEP World Conservation Monitoring legal status of any country, territory, city or Centre (UNEP-WCMC), based in Cambridge, area or its authority, or concerning the UK, is the specialist biodiversity information delimitation of its frontiers or boundaries. and assessment centre of the United Nations Environment Programme (UNEP), run cooperatively with WCMC, a UK charity. The © Copyright: 2010, European Commission Centre's mission is to evaluate and highlight the many values of biodiversity and put authoritative biodiversity knowledge at the centre of decision-making. Through the analysis and synthesis of global biodiversity knowledge the Centre provides authoritative, strategic and timely information for conventions, countries and organisations to use in the development and implementation of their policies and decisions. The UNEP-WCMC provides objective and scientifically rigorous procedures and services. -

Evaluation of 3 Milk Replacers for Hand-Rearing Indian Nilgai (Boselaphus Tragocamelus)
Evaluation of 3 Milk Replacers for Hand-Rearing Indian Nilgai (Boselaphus tragocamelus) Catherine D. Burch, M. Diehl, and M.S Edwards Zoological Society of San Diego, San Diego Wild Animal Park, San Diego, California Hoofstock neonates are often hand-raised for multiple reasons. Feeding maternal milk is optimal, but is rarely available for hand-rearing. Therefore, suitable substitutes must be identified. Published nutrient composition of maternal milk is frequently used as a guide. In many instances, however, no information is available regarding the milk composition of a particular species. Nilgai have been successfully raised at the Wild Animal Park using several formulas. It has yet to be determined which formula(s) produce the best response. Eight nilgai (3 females, 5 males) were received for hand-rearing and were randomly assigned 1 of the following 3 formulas for a blind study: evaporated cows (EC) :nonfat cows (NFC) 2: 1, EC: Milk Matrix 30/55®:water 10: 3: 12, or EC:SPF-Iac® 2: 1. EC: NFC 2: 1, which has been used for several seasons, was designated as the control. The composition of EC:30/55:H2O 10:3: 12 is comparable to the published composition of eland milk, a species taxonomically related to nilgai. EC:SPF-Iac® 2: 1 has lipid and carbohydrate composition intermediate to the other formulas. Response parameters monitored include weight gain, stool consistency, feeding response, and overall body condition. Based on the results, Indian nilgai can be hand-reared successfully on any of the 3 milk replacers. Of the 3, however, the group fed EC:NFC 2: 1 were the heaviest at weaning and the most vigorous throughout the hand-rearing process. -
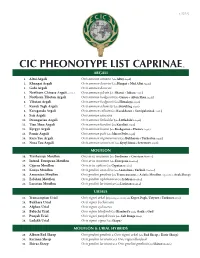
Cic Pheonotype List Caprinae©
v. 5.25.12 CIC PHEONOTYPE LIST CAPRINAE © ARGALI 1. Altai Argali Ovis ammon ammon (aka Altay Argali) 2. Khangai Argali Ovis ammon darwini (aka Hangai & Mid Altai Argali) 3. Gobi Argali Ovis ammon darwini 4. Northern Chinese Argali - extinct Ovis ammon jubata (aka Shansi & Jubata Argali) 5. Northern Tibetan Argali Ovis ammon hodgsonii (aka Gansu & Altun Shan Argali) 6. Tibetan Argali Ovis ammon hodgsonii (aka Himalaya Argali) 7. Kuruk Tagh Argali Ovis ammon adametzi (aka Kuruktag Argali) 8. Karaganda Argali Ovis ammon collium (aka Kazakhstan & Semipalatinsk Argali) 9. Sair Argali Ovis ammon sairensis 10. Dzungarian Argali Ovis ammon littledalei (aka Littledale’s Argali) 11. Tian Shan Argali Ovis ammon karelini (aka Karelini Argali) 12. Kyrgyz Argali Ovis ammon humei (aka Kashgarian & Hume’s Argali) 13. Pamir Argali Ovis ammon polii (aka Marco Polo Argali) 14. Kara Tau Argali Ovis ammon nigrimontana (aka Bukharan & Turkestan Argali) 15. Nura Tau Argali Ovis ammon severtzovi (aka Kyzyl Kum & Severtzov Argali) MOUFLON 16. Tyrrhenian Mouflon Ovis aries musimon (aka Sardinian & Corsican Mouflon) 17. Introd. European Mouflon Ovis aries musimon (aka European Mouflon) 18. Cyprus Mouflon Ovis aries ophion (aka Cyprian Mouflon) 19. Konya Mouflon Ovis gmelini anatolica (aka Anatolian & Turkish Mouflon) 20. Armenian Mouflon Ovis gmelini gmelinii (aka Transcaucasus or Asiatic Mouflon, regionally as Arak Sheep) 21. Esfahan Mouflon Ovis gmelini isphahanica (aka Isfahan Mouflon) 22. Larestan Mouflon Ovis gmelini laristanica (aka Laristan Mouflon) URIALS 23. Transcaspian Urial Ovis vignei arkal (Depending on locality aka Kopet Dagh, Ustyurt & Turkmen Urial) 24. Bukhara Urial Ovis vignei bocharensis 25. Afghan Urial Ovis vignei cycloceros 26. -

Chromosome Polymorphism in the Brazilian Dwarf Brocket Deer, Mazama Nana (Mammalia, Cervidae)
Genetics and Molecular Biology, 31, 1, 53-57 (2008) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Research Article Chromosome polymorphism in the Brazilian dwarf brocket deer, Mazama nana (Mammalia, Cervidae) Vanessa Veltrini Abril1,2 and José Maurício Barbanti Duarte2 1Programa de Pós-Graduação em Genética e Melhoramento Animal, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Campus de Jaboticabal, Jaboticabal, SP, Brazil. 2Núcleo de Pesquisa e Conservação de Cervídeos, Departamento de Zootecnia, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Campus de Jaboticabal, Jaboticabal, SP, Brazil. Abstract The Brazilian dwarf brocket deer (Mazama nana) is the smallest deer species in Brazil and is considered threatened due to the reduction and alteration of its habitat, the Atlantic Rainforest. Moreover, previous work suggested the pres- ence of intraspecific chromosome polymorphisms which may contribute to further population instability because of the reduced fertility arising from the deleterious effects of chromosome rearrangements during meiosis. We used G- and C-banding, and nucleolus organizer regions localization by silver-nitrate staining (Ag-NOR) to investigate the causes of this variation. Mazama nana exhibited eight different karyotypes (2n = 36 through 39 and FN = 58) result- ing from centric fusions and from inter and intraindividual variation in the number of B chromosomes (one to six). Most of the animals were heterozygous for a single fusion, suggesting one or several of the following: a) genetic in- stability in a species that has not reached its optimal karyotypic evolutionary state yet; b) negative selective pressure acting on accumulated rearrangements; and c) probable positive selection pressure for heterozygous individuals which maintains the polymorphism in the population (in contrast with the negative selection for many rearrangements within a single individual). -

Line Transect Surveys Underdetect Terrestrial Mammals: Implications for the Sustainability of Subsistence Hunting
RESEARCH ARTICLE Line Transect Surveys Underdetect Terrestrial Mammals: Implications for the Sustainability of Subsistence Hunting José M. V. Fragoso1☯¤*, Taal Levi2‡, Luiz F. B. Oliveira3‡, Jeffrey B. Luzar4‡, Han Overman5‡, Jane M. Read6‡, Kirsten M. Silvius7☯ 1 Stanford University, Stanford, CA, 94305–5020, United States of America, 2 Department of Fisheries and Wildlife, Oregon State University, Corvallis, OR, United States of America, 3 Departamento de Vertebrados, Museu Nacional, UFRJ, RJ, 20.940–040, Brazil, 4 Stanford University, Stanford, CA, 94305–5020, United States of America, 5 Environmental and Forest Biology, State University of New York-College of Environmental Science and Forestry, Syracuse, NY, 13210, United States of America, 6 Geography Department, Syracuse University, Syracuse, NY, 13244, United States of America, 7 Department of Forest Resources and Environmental Conservation, Virginia Tech, Blacksburg, VA, United States of America ☯ These authors contributed equally to this work. ¤ Current address: Biodiversity Science and Sustainability, California Academy of Sciences, San Francisco, California, United States of America OPEN ACCESS ‡ These authors contributed subsets of effort equally to this work. * [email protected] Citation: Fragoso JMV, Levi T, Oliveira LFB, Luzar JB, Overman H, Read JM, et al. (2016) Line Transect Surveys Underdetect Terrestrial Mammals: Implications for the Sustainability of Subsistence Abstract Hunting. PLoS ONE 11(4): e0152659. doi:10.1371/ journal.pone.0152659 Conservation of Neotropical game species must take into account the livelihood and food Editor: Mathew S. Crowther, University of Sydney, security needs of local human populations. Hunting management decisions should there- AUSTRALIA fore rely on abundance and distribution data that are as representative as possible of true Received: January 19, 2016 population sizes and dynamics. -

Biodiversity Assessment for Kyrgyzstan
Biodiversity Assessment for Kyrgyzstan Task Order under the Biodiversity & Sustainable Forestry IQC (BIOFOR) USAID CONTRACT NUMBER: LAG-I-00-99-00014-00 SUBMITTED TO: USAID CENTRAL ASIAN REPUBLICS MISSION, ALMATY, KAZAKHSTAN SUBMITTED BY: CHEMONICS INTERNATIONAL INC. WASHINGTON, D.C. JUNE 2001 TABLE OF CONTENTS SECTION I INTRODUCTION I-1 SECTION II STATUS OF BIODIVERSITY II-1 A. Overview II-1 B. Major Ecoregions II-1 C. Species Diversity II-3 D. Agrobiodiversity II-5 E. Threats to Biodiversity II-6 F. Resource Trends II-7 SECTION III STATUS OF BIODIVERSITY CONSERVATION III-1 A. Protected Areas III-1 B. Agriculture III-2 C. Forests III-2 D. Fisheries III-3 SECTION IV STRATEGIC AND POLICY FRAMEWORK IV-1 A. Institutional Framework IV-1 B. Legislative Framework IV-3 C. International Conventions and Agreements IV-5 D. Internationally Funded Programs IV-5 SECTION V SUMMARY OF FINDINGS V-1 SECTION VI RECOMMENDATIONS FOR IMPROVED BIODIVERSITY CONSERVATION VI-1 SECTION VII USAID/KYRGYZSTAN VII-1 A. Impact of USAID Program on Biodiversity VII-1 B. Recommendations VII-1 ANNEX A SECTIONS 117 AND 119 OF THE FOREIGN ASSISTANCE ACT A-1 ANNEX B SCOPE OF WORK B-1 ANNEX C LIST OF PERSONS CONTACTED C-1 ANNEX D LISTS OF RARE AND ENDANGERED SPECIES OF KYRGYZSTAN D-1 ANNEX E MAP OF ECOSYSTEMS AND PROTECTED AREAS OF KYRGYZSTAN E-1 ANNEX F PROTECTED AREAS IN KYRGYZSTAN F-1 ANNEX G SCHEDULE OF TEAM VISITS G-1 ANNEX H INSTITUTIONAL CONSTRAINTS AND OPPORTUNITIES (FROM NBSAP) H-1 ANNEX I CENTRAL ASIA TRANSBOUNDARY BIODIVERSITY PROJECT I-1 ACRONYMS BEO Bureau Environmental Officer BIOFOR Biodiversity and Sustainable Forestry BSAP Biodiversity Strategy and Action Plan CAR Central Asian Republics CITES Convention on International Trade in Endangered Species CTO Contracting Technical Officer DC District of Columbia EE Europe and Eurasia FAA Foreign Assistance Act GEF Global Environment Fund GIS Geographic Information Systems GTZ German Agency for Technical Cooperation ha hectare I.A. -

Whole-Genome Sequencing of Wild Siberian Musk
Yi et al. BMC Genomics (2020) 21:108 https://doi.org/10.1186/s12864-020-6495-2 RESEARCH ARTICLE Open Access Whole-genome sequencing of wild Siberian musk deer (Moschus moschiferus) provides insights into its genetic features Li Yi1†, Menggen Dalai2*†, Rina Su1†, Weili Lin3, Myagmarsuren Erdenedalai4, Batkhuu Luvsantseren4, Chimedragchaa Chimedtseren4*, Zhen Wang3* and Surong Hasi1* Abstract Background: Siberian musk deer, one of the seven species, is distributed in coniferous forests of Asia. Worldwide, the population size of Siberian musk deer is threatened by severe illegal poaching for commercially valuable musk and meat, habitat losses, and forest fire. At present, this species is categorized as Vulnerable on the IUCN Red List. However, the genetic information of Siberian musk deer is largely unexplored. Results: Here, we produced 3.10 Gb draft assembly of wild Siberian musk deer with a contig N50 of 29,145 bp and a scaffold N50 of 7,955,248 bp. We annotated 19,363 protein-coding genes and estimated 44.44% of the genome to be repetitive. Our phylogenetic analysis reveals that wild Siberian musk deer is closer to Bovidae than to Cervidae. Comparative analyses showed that the genetic features of Siberian musk deer adapted in cold and high-altitude environments. We sequenced two additional genomes of Siberian musk deer constructed demographic history indicated that changes in effective population size corresponded with recent glacial epochs. Finally, we identified several candidate genes that may play a role in the musk secretion based on transcriptome analysis. Conclusions: Here, we present a high-quality draft genome of wild Siberian musk deer, which will provide a valuable genetic resource for further investigations of this economically important musk deer.