Chapter 5 Medical Classifications and Terminologies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MHSUDS Information Notice No.: 16-051
State of California—Health and Human Services Agency Department of Health Care Services JENNIFER KENT EDMUND G. BROWN JR. DIRECTOR GOVERNOR DATE: October 7, 2016 MHSUDS INFORMATION NOTICE N O.: 16-051 TO: COUNTY BEHAVIORAL HEALTH DIRECTORS COUNTY BEHAVIORAL HEALTH DIRECTORS ASSOCIATION OF CALIFORNIA CALIFORNIA COUNCIL OF COMMUNITY BEHAVIORAL HEALTH AGENCIES CALIFORNIA ALLIANCE OF CHILD AND FAMILY SERVICES SUBJECT: IMPLEMENTATION OF THE DIAGNOSTIC AND STATISTICAL MANUAL OF MENTAL DISORDERS (DSM-5), FIFTH EDITION REFERENCES: MHSD INFORMATION NOTICE 13-22 MHSUDS INFORMATION NOTICE 14-040 MHSUDS INFORMATION NOTICE 15-003 MHSUDS INFORMATION NOTICE 15-030 PURPOSE The purpose of this Information Notice is to direct Mental Health Plans (MHPs) to use the American Psychiatric Association (APA) DS M-5 to make diagnostic determinations for the purposes of determining if beneficiaries meet medical necessity criteria for Medi-Cal specialty mental health services (SMHS). This Information Notice updates information previously provided in Mental Health Services Division (MHSD) Information Notice 13-22. BACKGROUND The APA issued the DSM-5 on May 20, 2013. The DSM-5 provides diagnostic criteria and codes as well as corresponding International Classification of Diseases-10 (ICD-10) codes. Since March 2013, the Department of Health Care Services (DHCS) has collaborated with internal and external stakeholders and partners to address implementation issues specific to SMHS related to the DSM-5. Mental Health & Substance Use Disorder Services 1501 Capitol Avenue, MS 4000, P.O. Box 997413 Sacramento, CA 95899-7413 Phone: (916) 440-7800 Fax: (916) 319-8219 Internet Address: www.dhcs.ca.gov MHSUDS INFORMATION NOTICE N O.: 16-051 October 7, 2016 Page 2 DHCS has issued previous MHSUDS Information Notices1 that instruct MHPs to report only ICD-10 codes for claiming and diagnoses reporting purposes and provide ICD-10 procedural and diagnosis crosswalk documents f or SMHS. -

A Reason for Visit Classification for Ambulatory Care
DATA EVALUATION AND METHODS RESEARCH Series 2 Number 78 A Reasonfor Visit Classification for AmbulatoryCare This report presents a classification system developed to code rea- sons for seeking ambulatory medical care. DHEW Publication No. (PHS) 79-1352 U.S. DEPARTMENT OF HEALTH, EDUCATION, AND WELFARE Public Health Service Office of the Assistant Secretary for Health National Center for Health Statistics Hyattsville, Md. February 1979 Library of Congress Cataloging in Publication Data Schneider, Don. A reason for visit classification for ambulatory care. (Vital and health statistics: Series 2, Data evaluation and methods research; no. 78) (DHEW publication; no. (PHS) 79-1352) Includes bibliographical references and index. 1. Nosology. 2. Ambulatory medical care. I. Appleton, Linda, joint author. II. McLemore, Thomas, ,joint author. III. Title. IV. Series: United States. National Center for Health Sta- tistics. Vital and health statistics: Series 2, Data evaluation and methods research; no. 78. V. Series: United States. Dept. of Health, Education and Welfare. DHEW publication; no. (PHS) 79-1352. [DNLM: 1. Ambulatory care. 2. Classification. W2 A N148vb no. 78] RA409.U45 no. 78 [RB115] 312’.07’23s [616’.001’2] 78-11549 NATIONAL CENTER FOR HEALTH STATISTICS DOROTHY P. RICE, Director ROBERT A. ISRAEL, Deputy Director JACOB J. FELDMAN, Ph.D., Associate Director for Analysis GAIL F. FISHER, Ph.D., Associate Director for the Cooperative Health Statistics System ELIJAH L. WHITE, Associate Director for Data Systems JAMES T. BAIRD, JR., Ph.D., Associate .Directorfor International Statistics ROBERT C, HUBER, Associate Director for Management MONROE G. SIRKEN, Ph. D., Associate Director for Mathematiccd Statistics PETER L. HURLEY, Associate Director for Operations JAMES M. -

ICD-10 International Statistical Classification of Diseases and Related Health Problems
ICD-10 International Statistical Classification of Diseases and Related Health Problems 10th Revision Volume 2 Instruction manual 2010 Edition WHO Library Cataloguing-in-Publication Data International statistical classification of diseases and related health problems. - 10th revision, edition 2010. 3 v. Contents: v. 1. Tabular list – v. 2. Instruction manual – v. 3. Alphabetical index. 1.Diseases - classification. 2.Classification. 3.Manuals. I.World Health Organization. II.ICD-10. ISBN 978 92 4 154834 2 (NLM classification: WB 15) © World Health Organization 2011 All rights reserved. Publications of the World Health Organization are available on the WHO web site (www.who.int) or can be purchased from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press through the WHO web site (http://www.who.int/about/licensing/copyright_form). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. -

Clinical Vocabularies for Global Real World Evidence (RWE) | Evidera
Clinical Vocabularies for Global RWE Analysis Don O’Hara, MS Senior Research Associate, Real-World Evidence, Evidera Vernon F. Schabert, PhD Senior Research Scientist, Real-World Evidence, Evidera Introduction significant volume of real-world evidence (RWE) analyses continue to be conducted with data A repurposed from healthcare administrative Don O’Hara Vernon F. Schabert databases. The range of sources represented by those databases has grown in response to demand for richer description of patient health status and outcomes. information for improved quality and coordination Data availability, including the range of available data of care, often using standardized messaging systems sources, has grown unevenly across the globe in response such as Health Level 7 (HL7). These messages are to country-specific market and regulatory dynamics. only as good as the standardization of codes between Nonetheless, as demand globalizes for RWE insights from message senders and receivers, which motivates the databases, those demands increase pressure on analysts encoding of facts using common code sets. The second to find ways to bridge differences between local data is the increased availability of common data models to sources to achieve comparable insights across regions. standardize the extraction and analysis of these data for RWE and drug safety purposes. While common data One of the challenges in bridging differences across models make compromises on the structure of tables databases is the codes used to represent key clinical and fields extracted from healthcare systems such as facts. Historically, RWE database studies have leveraged electronic medical records (EMR) and billing systems, local code sets for cost-bearing healthcare services they can improve consistency and replicability of analyses such as drugs, procedures, and laboratory tests. -
Non-Interventional (Ni) Study Report
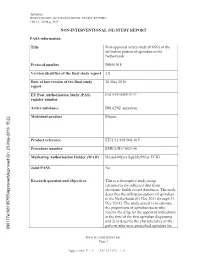
Apixaban B0661018 NON-INTERVENTIONAL STUDY REPORT FINAL, 20 May 2015 NON-INTERVENTIONAL (NI) STUDY REPORT PASS information Title Post-approval safety study (PASS) of the utilization pattern of apixaban in the Netherlands Protocol number B0661018 Version identifier of the final study report 1.0 Date of last version of the final study 20 May 2016 report EU Post Authorisation Study (PAS) ENCEPP/SDPP/5177 register number Active substance B01AF02 apixaban Medicinal product Eliquis -2016 15:22 y Product reference EU/1/11/691/001-015 Procedure number EMEA/H/C/002148 Marketing Authorisation Holder (MAH) Bristol-Myers Squibb/Pfizer EEIG roved On: 23-Ma Joint PASS No pp Research question and objectives This is a descriptive study using roved\A retrospectively collected data from pp electronic health record databases. The study describes the utilization pattern of apixaban in the Netherlands (01 Dec 2011 through 31 Dec 2014). The study aimed 1) to estimate the proportions of apixaban users who receive the drug for the approved indications at the time of the first apixaban dispensing and 2) to describe the characteristics of the patients who were prescribed apixaban for 090177e188105755\A PFIZER CONFIDENTIAL Page 1 Approved v 1.0 930102376 1.0 Apixaban B0661018 NON-INTERVENTIONAL STUDY REPORT FINAL, 20 May 2015 on-label and off-label indications. Country(-ies) of study The Netherlands Author Marketing Authorisation Holder(s) Marketing Authorisation Holder(s) Bristol-Myers Squibb/Pfizer EEIG -2016 15:22 y MAH contact person roved On: 23-Ma pp roved\A pp This document contains confidential information belonging to Pfizer. -

International Classification of Diseases- 10Th Revision (ICD-10)
Massachusetts Deaths 1999 Frequently Asked Questions: International Classification of Diseases- 10th Revision (ICD-10) Effective with data year 1999, the International Classification of Diseases, Tenth Revision (ICD-10) is used to code and classify causes of death. This change in coding systems will affect the comparison of mortality data between 1999 and previous years. Definitions and Concepts What is ICD-10? ICD-10 is an abbreviation for the International Classification of Diseases- Tenth revision. The International Classification of Diseases is a classification system developed by the World Health Organization (WHO). The United States uses the ICD in accordance with an international agreement. The purpose of an international classification system is to promote international comparability in collecting, classifying, and tabulating mortality statistics. Why has the ICD been revised? The ICD is revised to reflect changes in medical classification practices. The ICD was first implemented in 1900, and has undergone revisions approximately every ten years, except for the Ninth revision which was in effect between 1979-1998. There has been an increase in the number of specific cause of death codes from about 4,000 codes in ICD-9 to about 8,000 codes in ICD-10. Beginning with 1999, mortality data are coded according to the Tenth revision of the ICD. Can I compare data classified in ICD-10 to data classified in ICD-9? Differences in the coding between ICD-9 and ICD-10 make direct comparisons between the two classification systems difficult for three reasons: 1) there have been changes made in the codes that are assigned to causes of death; 2) there have been changes to the rules used to determine the underlying cause of death; and 3) there have been changes in the codes that comprise the leading cause of death categories. -

(Child Core Set) Technical Specifications and Reso
Core Set of Children's Health Care Quality Measures for Medicaid and CHIP (Child Core Set) Technical Specifications and Resource Manual for Federal Fiscal Year 2019 Reporting February 2019 Center for Medicaid and CHIP Services Centers for Medicare & Medicaid Services This page left blank for double-sided copying ii ACKNOWLEDGMENTS For Proprietary Codes: CPT® codes copyright 2018 American Medical Association. All rights reserved. CPT is a trademark of the American Medical Association. No fee schedules, basic units, relative values or related listings are included in CPT. The AMA assumes no liability for the data contained herein. Applicable FARS/DFARS restrictions apply to government use. SNOMED CLINICAL TERMS (SNOMED CT®) copyright 2004-2018 The International Health Terminology Standards Development Organisation. All Rights Reserved. The International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) is published by the World Health Organization (WHO). ICD-9-CM is an official Health Insurance Portability and Accountability Act standard. The International Classification of Diseases, 9th Revision, Procedure Coding System (ICD-9-PCS) is published by the World Health Organization (WHO). ICD-9-PCS is an official Health Insurance Portability and Accountability Act standard. The International Classification of Diseases, 10th Revision, Clinical Modification (ICD-10-CM) is published by the World Health Organization (WHO). ICD-10-CM is an official Health Insurance Portability and Accountability Act standard. The International Classification of Diseases, 10th Revision, Procedure Coding System (ICD-10-PCS) is published by the World Health Organization (WHO). ICD-10-PCS is an official Health Insurance Portability and Accountability Act standard. This material contains content from LOINC® (http://loinc.org). -

Inappropriate Primary Diagnosis Codes Policy, Professional
Commercial Reimbursement Policy CMS 1500 Policy Number 2020R0122C Inappropriate Primary Diagnosis Codes Policy, Professional IMPORTANT NOTE ABOUT THIS REIMBURSEMENT POLICY You are responsible for submission of accurate claims. This reimbursement policy is intended to ensure that you are reimbursed based on the code or codes that correctly describe the health care services provided. UnitedHealthcare reimbursement policies may use Current Procedural Terminology (CPT®*), Centers for Medicare and Medicaid Services (CMS) or other coding guidelines. References to CPT or other sources are for definitional purposes only and do not imply any right to reimbursement. This reimbursement policy applies to all health care services billed on CMS 1500 forms and, when specified, to those billed on UB04 forms. Coding methodology, industry-standard reimbursement logic, regulatory requirements, benefits design and other factors are considered in developing reimbursement policy. This information is intended to serve only as a general reference resource regarding UnitedHealthcare’s reimbursement policy for the services described and is not intended to address every aspect of a reimbursement situation. Accordingly, UnitedHealthcare may use reasonable discretion in interpreting and applying this policy to health care services provided in a particular case. Further, the policy does not address all issues related to reimbursement for health care services provided to UnitedHealthcare enrollees. Other factors affecting reimbursement may supplement, modify or, in some cases, supersede this policy. These factors may include, but are not limited to: legislative mandates, the physician or other provider contracts, the enrollee’s benefit coverage documents and/or other reimbursement, medical or drug policies. Finally, this policy may not be implemented exactly the same way on the different electronic claims processing systems used by UnitedHealthcare due to programming or other constraints; however, UnitedHealthcare strives to minimize these variations. -

G11.11: New Icd-10-Cm (Diagnosis) Code for Friedreich's Ataxia
G11.11: NEW ICD-10-CM (DIAGNOSIS) CODE FOR FRIEDREICH’S ATAXIA EFFECTIVE OCTOBER 1, 2020 CALL TO ACTION – PROVIDE THIS INFO TO YOUR CLINICIANS Starting on October 1, 2020, at your next appointments with any clinician, ask that your ICD-10 diagnosis code for FA be updated to G11.11 (previous code for FA was G11.1 > add a “1” > G11.11). Your clinician can use multiple ICD-10 diagnosis codes based on your need for medical care (e.g. G11.11 – FA, I49.9 - cardiac arrhythmia, Z13.1 – encounter for screening for diabetes mellitus). Please email [email protected] if you or your clinician has questions regarding use of G11.11 PREVIOUS CODING FOR FRIEDREICH’S ATAXIA ICD-9 system had a specific code for FA: 334.0 In conversion to ICD-10, the code for FA became less specific because it covered a group of conditions described as “early-onset cerebellar ataxias”. The previous FA ICD-10 diagnosis code was G11.1. BENEFITS OF THE NEW FA-SPECIFIC ICD-10-CM CODE G11.11 • Fewer rejected health insurance claims • Payer coverage determinations without need for direct access to patients’ medical records for review of medical necessity • Monitor adherence to clinical management guidelines and track care outcomes • Evaluate symptom progression of FA; additional ICD-10 codes can be applied to reflect context of FA diagnosis, such as for falls, surgeries, medications, and co-morbidities • Enhanced ability to track disease prevalence BACKGROUND ON ICD CODING SYSTEM ICD = International Classification of Diseases The ICD coding system was established by the World Health Organization; currently used by ~100 countries for reporting cause of death and epidemiology statistics. -

ICF: REPRESENTING the PATIENT BEYOND a MEDICAL CLASSIFICATION of DIAGNOSES Posted on July 17, 2009 by Administrator
ICF: REPRESENTING THE PATIENT BEYOND A MEDICAL CLASSIFICATION OF DIAGNOSES Posted on July 17, 2009 by Administrator Category: Clinical Terms & Vocabularies Tag: ICF Page: 1 by Kathy Giannangelo, RHIA, CCS; Sue Bowman, RHIA, CCS; Michelle Dougherty, RHIA, CHP; and Susan Fenton, MBA, RHIA Abstract The International Classification of Functioning, Disability and Health (ICF) is a component essential to ensuring the collection of accurate and complete healthcare data that correctly reflect the care provided to individuals. In fact, many countries outside of the United States have found uses for ICF. While research continues in the United States on the potential value of implementing ICF, deliberations are establishing the need to implement ICF to develop knowledge about the physical, mental, and social functioning of patients. In the course of these deliberations, issues related to current data collection activities, the use of ICF in an electronic health record (EHR) system, training requirements, and terminology maps are beginning to emerge. Introduction The International Classification of Functioning, Disability and Health (ICF) is one of the World Health Organization’s Family of International Classifications (WHO-FIC).1 The purpose of WHO-FIC is to promote the appropriate selection of classifications in the range of settings in the healthcare field around the world. Since a single classification system cannot encompass all types of healthcare information or provide the level of detail desired for various uses of healthcare data, multiple classifications have been developed to meet specific user requirements. WHO-FIC provides a framework to code a wide range of information about health (for example, diagnosis, functioning, disability, and reasons for contact with health services) and uses standardized language, permitting communication about health and healthcare around the world and among various disciplines and sciences. -

ROR*1.5*36 User Manual
Clinical Case Registries (CCR) Version 1.5 User Manual Documentation Revised May 2020 For Patch ROR*1.5*36 Department of Veterans Affairs Office of Enterprise Development Health Data Systems – Registries THIS PAGE INTENTIONALLY LEFT BLANK Revision History Date Description Author Role May, 2020 Final release for Patch ROR*1.5*36. See Table REDACTED Project Manager 54Table 52 for Details. M Developer M Developer Software QA Analyst Software QA Analyst Delphi Developer November, Final release for Patch ROR*1.5*35. See Table REDACTED Project Manager 2019 52Table 52 for Details. M Developer M Developer Software QA Analyst Delphi Developer March, 2019 Final release for Patch ROR*1.5*34. See Table REDACTED Project Manager 50 for Details. M Developer M Developer Software QA Analyst Delphi Developer July, 2018 Final release for Patch ROR*1.5*33. See Table REDACTED Project Manager 48Table 52 for Details. M Developer M Developer Software QA Analyst Delphi Developer April, 2018 Final release for Patch ROR*1.5*32. See Table REDACTED Project Manager 46Error! Reference source not found. for Details. M Developer Software QA Analyst Delphi Developer May, 2020 Clinical Case Registries ROR*1.5*36 i User Manual Date Description Author Role November, Final release for Patch ROR*1.5*31. See Table REDACTED Project Manager 2017 44 for Details. M Developer M Developer Software QA Analyst Software QA Analyst Delphi Developer May, 2017 Final release for Patch ROR*1.5*30. See Table REDACTED Project Manager 42 for Details. M Developer SQA Analyst Delphi Developer December, Final release for Patch ROR*1.5*29. -

Toward a Preliminary Layered Network Model Representation of Medical Coding in Clinical Decision Support Systems
146 Int'l Conf. Health Informatics and Medical Systems | HIMS'15 | Toward a Preliminary Layered Network Model Representation of Medical Coding in Clinical Decision Support Systems H. Mallah1, I. Zaarour2, A. Kalakech2 1 Doctoral School of Management Information Systems, Lebanese University, Beirut, Lebanon 2 Management Information Systems Department, Lebanese University, Beirut, Lebanon Abstract - The aim of this research is to describe the impact increase in medical errors, poor trainings to human resources of Medical Codes (MC) on building a successful Clinical and their derivatives. Many studies were done on how to Decision Support System (CDSS) that helps in structuring a computerize medical processes, especially critical decisions beneficial Information Model, we highlight a preliminary in medical fields, using Artificial Intelligence. From these Architectural Layered network model to assist health care needs, Clinical Decision Support System (CDSS) and givers and researchers in representing their work in a Medical classification or medical coding gave birth after the unified manner. Via a descriptive methodology, we development of the Electronic Health Record (EHR) and the incorporate the relationship between (MC) and (CDSS), we Computer-based Physician Order Entry (CPOE). CDSS is discussed a brief state of art of (CDSS) as well as medical the key of success of clinical information systems designed coding as a process of converting descriptions of to assist health care providers in decision making during the medical diagnoses and procedures into universal medical process of care. This means that a clinician would cooperate codes and numbers. Integrated with CDSS, medical codes with a CDSS to help determine diagnosis, analysis of patient are illustrated as an advanced search engine in the data.