Sequence Variations of ACVRL1 Play a Critical Role in Hepatic Vascular
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of Pulmonary Arteriovenous Malformations in ACVRL1 Versus ENG Mutation Carriers in Hereditary Hemorrhagic Telangiectasia
© American College of Medical Genetics and Genomics ORIGINAL RESEARCH ARTICLE Characterization of pulmonary arteriovenous malformations in ACVRL1 versus ENG mutation carriers in hereditary hemorrhagic telangiectasia Weiyi Mu, ScM1, Zachary A. Cordner, MD, PhD2, Kevin Yuqi Wang, MD3, Kate Reed, MPH, ScM4, Gina Robinson, RN5, Sally Mitchell, MD5 and Doris Lin, MD, PhD5 Purpose: Pulmonary arteriovenous malformations (pAVMs) are mutation carriers to have pAVMs (P o 0.001) or multiple lesions major contributors to morbidity and mortality in hereditary (P = 0.03), and to undergo procedural intervention (P = 0.02). hemorrhagic telangiectasia (HHT). Mutations in ENG and ACVRL1 Additionally, pAVMs in ENG carriers were more likely to exhibit underlie the vast majority of clinically diagnosed cases. The aims of bilateral lung involvement and growth over time, although this did this study were to characterize and compare the clinical and not reach statistical significance. The HHT severity score was morphologic features of pAVMs between these two genotype significantly higher in ENG than in ACVRL1 (P = 0.02). groups. Conclusion: The propensity and multiplicity of ENG-associated Methods: Sixty-six patients with HHT and affected family pAVMs may contribute to the higher disease severity in this members were included. Genotype, phenotypic data, and imaging genotype, as reflected by the HHT severity score and the frequency were obtained from medical records. Morphologic features of of interventional procedures. pAVMs were analyzed using computed tomography angiography. Genet Med HHT symptoms, pAVM imaging characteristics, frequency of advance online publication 19 October 2017 procedural intervention, and HHT severity scores were compared Key Words: ACVRL1; ENG; genotype-phenotype correlation; between ENG and ACVRL1 genotype groups. -

Hereditary Hemorrhagic Telangiectasia: Diagnosis and Management From
REVIEW ARTICLE Hereditary hemorrhagic telangiectasia: Ferrata Storti diagnosis and management from Foundation the hematologist’s perspective Athena Kritharis,1 Hanny Al-Samkari2 and David J Kuter2 1Division of Blood Disorders, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ and 2Hematology Division, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA ABSTRACT Haematologica 2018 Volume 103(9):1433-1443 ereditary hemorrhagic telangiectasia (HHT), also known as Osler- Weber-Rendu syndrome, is an autosomal dominant disorder that Hcauses abnormal blood vessel formation. The diagnosis of hered- itary hemorrhagic telangiectasia is clinical, based on the Curaçao criteria. Genetic mutations that have been identified include ENG, ACVRL1/ALK1, and MADH4/SMAD4, among others. Patients with HHT may have telangiectasias and arteriovenous malformations in various organs and suffer from many complications including bleeding, anemia, iron deficiency, and high-output heart failure. Families with the same mutation exhibit considerable phenotypic variation. Optimal treatment is best delivered via a multidisciplinary approach with appropriate diag- nosis, screening and local and/or systemic management of lesions. Antiangiogenic agents such as bevacizumab have emerged as a promis- ing systemic therapy in reducing bleeding complications but are not cur- ative. Other pharmacological agents include iron supplementation, antifibrinolytics and hormonal treatment. This review discusses the biol- ogy of HHT, management issues that face -

Activin Receptor Type IA (ACVR1) Antibody (Center N153) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP7101A
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Activin Receptor Type IA (ACVR1) Antibody (Center N153) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP7101A Specification Activin Receptor Type IA (ACVR1) Antibody (Center N153) - Product Information Application WB, IHC-P,E Primary Accession Q04771 Other Accession P80201, P37172, Q28041 Reactivity Human Predicted Bovine, Mouse, Rat Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Antigen Region 138-170 Activin Receptor Type IA (ACVR1) Antibody Western blot analysis of ACVR1 (arrow) using (Center N153) - Additional Information rabbit polyclonal ACVR1 Antibody (Center N153) (Cat.#AP7101a). 293 cell lysates (2 Gene ID 90 ug/lane) either nontransfected (Lane 1) or transiently transfected with the ACVR1 gene Other Names (Lane 2) (Origene Technologies). Activin receptor type-1, Activin receptor type I, ACTR-I, Activin receptor-like kinase 2, ALK-2, Serine/threonine-protein kinase receptor R1, SKR1, TGF-B superfamily receptor type I, TSR-I, ACVR1, ACVRLK2 Target/Specificity This Activin Receptor Type IA (ACVR1) antibody is generated from rabbits immunized with a KLH conjugated synthetic peptide between 138-170 amino acids from the Central region of human Activin Receptor Type IA (ACVR1). Dilution WB~~1:1000 ACVR1 Antibody (Cat. #AP7101a) western IHC-P~~1:50~100 blot analysis in HL-60 cell line lysates (35ug/lane).This demonstrates the ACVR1 Format antibody detected the ACVR1 protein (arrow). Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is prepared by Saturated Ammonium Sulfate (SAS) precipitation followed by dialysis against PBS. Storage Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Maintain refrigerated at 2-8°C for up to 2 weeks. -

ALK2 / ACVR1 Antibody (Internal) Goat Polyclonal Antibody Catalog # ALS12616
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 ALK2 / ACVR1 Antibody (Internal) Goat Polyclonal Antibody Catalog # ALS12616 Specification ALK2 / ACVR1 Antibody (Internal) - Product Information Application WB Primary Accession Q04771 Reactivity Human, Mouse, Rat, Rabbit, Monkey, Pig, Horse, Bovine, Dog Host Goat Clonality Polyclonal Calculated MW 57kDa KDa ALK2 / ACVR1 Antibody (Internal) - Additional Antibody (0.3 ug/ml) staining of Human Information Umbilical Cord lysate (35 ug protein in RIPA buffer). Gene ID 90 Other Names ALK2 / ACVR1 Antibody (Internal) - Activin receptor type-1, 2.7.11.30, Activin Background receptor type I, ACTR-I, Activin receptor-like kinase 2, ALK-2, Serine/threonine-protein On ligand binding, forms a receptor complex kinase receptor R1, SKR1, TGF-B consisting of two type II and two type I superfamily receptor type I, TSR-I, ACVR1, transmembrane serine/threonine kinases. Type ACVRLK2 II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind Target/Specificity and activate SMAD transcriptional regulators. Human ACVR1 / ALK2. Receptor for activin. May be involved for left-right pattern formation during Reconstitution & Storage embryogenesis (By similarity). Store at -20°C. Minimize freezing and thawing. ALK2 / ACVR1 Antibody (Internal) - References Precautions ALK2 / ACVR1 Antibody (Internal) is for Matsuzaki K.,et al.J. Biol. Chem. research use only and not for use in diagnostic or therapeutic procedures. 268:12719-12723(1993). ten Dijke P.,et al.Oncogene 8:2879-2887(1993). Oppermann F.S.,et al.Mol. Cell. Proteomics ALK2 / ACVR1 Antibody (Internal) - Protein Information 8:1751-1764(2009). Umasankar P.K.,et al.Nat. -

Context-Dependent Roles in Cell and Tissue Physiology
Downloaded from http://cshperspectives.cshlp.org/ on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press TGF-b and the TGF-b Family: Context-Dependent Roles in Cell and Tissue Physiology Masato Morikawa,1 Rik Derynck,2 and Kohei Miyazono3 1Ludwig Cancer Research, Science for Life Laboratory, Uppsala University, Biomedical Center, SE-751 24 Uppsala, Sweden 2Department of Cell and Tissue Biology, University of California at San Francisco, San Francisco, California 94143 3Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan Correspondence: [email protected] The transforming growth factor-b (TGF-b) is the prototype of the TGF-b family of growth and differentiation factors, which is encoded by 33 genes in mammals and comprises homo- and heterodimers. This review introduces the reader to the TGF-b family with its complexity of names and biological activities. It also introduces TGF-b as the best-studied factor among the TGF-b family proteins, with its diversity of roles in the control of cell proliferation and differentiation, wound healing and immune system, and its key roles in pathology, for exam- ple, skeletal diseases, fibrosis, and cancer. lthough initially thought to stimulate cell TGF-b has been well documented in most cell Aproliferation, just like many growth factors, types, and has been best characterized in epithe- it became rapidly accepted that transforming lial cells. The bifunctional and context-depen- growth factor b (TGF-b) is a bifunctional reg- dent nature of TGF-b activities was further con- ulator that either inhibits or stimulates cell pro- firmed in a large variety of cell systems and liferation. -

Primary Chicken and Duck Endothelial Cells Display a Differential Response to Infection with Highly Pathogenic Avian Influenza Virus
G C A T T A C G G C A T genes Article Primary Chicken and Duck Endothelial Cells Display a Differential Response to Infection with Highly Pathogenic Avian Influenza Virus Zhen Wei Marcus Tong 1,†, Anjana C. Karawita 1,2,† , Colin Kern 3, Huaijun Zhou 3 , Jane E. Sinclair 1 , Limin Yan 1, Keng Yih Chew 1, Sue Lowther 2, Lee Trinidad 2, Arjun Challagulla 2 , Karel A. Schat 4 , Michelle L. Baker 2 and Kirsty R. Short 1,5,* 1 School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane 4072, Australia; [email protected] (Z.W.M.T.); [email protected] (A.C.K.); [email protected] (J.E.S.); [email protected] (L.Y.); [email protected] (K.Y.C.) 2 CSIRO, Australian Centre for Disease Preparedness, Health, and Biosecurity Business Unit, Geelong 3219, Australia; [email protected] (S.L.); [email protected] (L.T.); [email protected] (A.C.); [email protected] (M.L.B.) 3 Department of Animal Science, University of California, Davis, CA 95616, USA; [email protected] (C.K.); [email protected] (H.Z.) 4 Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA; [email protected] 5 Australian Infectious Diseases Research Centre, The University of Queensland, Brisbane 4072, Australia * Correspondence: [email protected] † These authors contributed equally to this work. Citation: Tong, Z.W.M.; Karawita, A.C.; Kern, C.; Zhou, H.; Sinclair, J.E.; Abstract: Highly pathogenic avian influenza viruses (HPAIVs) in gallinaceous poultry are associated Yan, L.; Chew, K.Y.; Lowther, S.; Trinidad, L.; Challagulla, A.; et al. -

Tgfβ/BMP Signaling Pathway in Cartilage Homeostasis
cells Review TGFβ/BMP Signaling Pathway in Cartilage Homeostasis Nathalie G.M. Thielen , Peter M. van der Kraan and Arjan P.M. van Caam * Experimental Rheumatology, Radboud University Medical Center, Geert Grooteplein 28, 6525 GA Nijmegen, The Netherlands * Correspondence: [email protected]; Tel.: +31-24-10513; Fax: +31-24-3540403 Received: 2 July 2019; Accepted: 19 August 2019; Published: 24 August 2019 Abstract: Cartilage homeostasis is governed by articular chondrocytes via their ability to modulate extracellular matrix production and degradation. In turn, chondrocyte activity is regulated by growth factors such as those of the transforming growth factor β (TGFβ) family. Members of this family include the TGFβs, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs). Signaling by this protein family uniquely activates SMAD-dependent signaling and transcription but also activates SMAD-independent signaling via MAPKs such as ERK and TAK1. This review will address the pivotal role of the TGFβ family in cartilage biology by listing several TGFβ family members and describing their signaling and importance for cartilage maintenance. In addition, it is discussed how (pathological) processes such as aging, mechanical stress, and inflammation contribute to altered TGFβ family signaling, leading to disturbed cartilage metabolism and disease. Keywords: transforming growth factor β; bone morphogenetic proteins; osteoarthritis; cartilage; SMADs; aging; joint loading; inflammation; linker modifications 1. Introduction The transforming growth factor β (TGFβ) family of polypeptide growth factors controls development and homeostasis of many tissues, including articular cartilage. Articular cartilage is the connective tissue covering joint surfaces and is a type of hyaline cartilage. This tissue is key in facilitating movement with its smooth lubricated surface, and it functions as a shock absorber to disperse forces acting upon movement with its physical properties. -

Signalling Pathways Regulating Organ-Specific Endothelial
REVIEWS The quiescent endothelium: signalling pathways regulating organ- specific endothelial normalcy Nicolas Ricard 1, Sabine Bailly2, Christophe Guignabert 3,4 and Michael Simons 1,5 ✉ Abstract | Endothelial cells are at the interface between circulating blood and tissues. This position confers on them a crucial role in controlling oxygen and nutrient exchange and cellular trafficking between blood and the perfused organs. The endothelium adopts a structure that is specific to the needs and function of each tissue and organ and is subject to tissue-specific signalling input. In adults, endothelial cells are quiescent, meaning that they are not proliferating. Quiescence was considered to be a state in which endothelial cells are not stimulated but are instead slumbering and awaiting activating signals. However, new evidence shows that quiescent endothelium is fully awake, that it constantly receives and initiates functionally important signalling inputs and that this state is actively regulated. Signalling pathways involved in the maintenance of functionally quiescent endothelia are starting to be identified and are a combination of endocrine, autocrine, paracrine and mechanical inputs. The paracrine pathways confer a microenvironment on the endothelial cells that is specific to the perfused organs and tissues. In this Review, we present the current knowledge of organ-specific signalling pathways involved in the maintenance of endothelial quiescence and the pathologies associated with their disruption. Linking organ- specific pathways -

TGF-Β Prodomain Alignments Reveal Unexpected Cysteine Conservation Consistent with Phylogenetic Predictions of Cross-Subfamily
| INVESTIGATION TGF-b Prodomain Alignments Reveal Unexpected Cysteine Conservation Consistent with Phylogenetic Predictions of Cross-Subfamily Heterodimerization Robert G. Wisotzkey* and Stuart J. Newfeld†,1 *Sema4 Genomics, Stamford, Connecticut 06902 and †School of Life Sciences, Arizona State University, Tempe, Arizona 85287-4501 ORCID ID: 0000-0003-1400-7978 (S.J.N.) ABSTRACT Evolutionary relationships between prodomains in the TGF-b family have gone unanalyzed due to a perceived lack of conservation. We developed a novel approach, identified these relationships, and suggest hypotheses for new regulatory mechanisms in TGF-b signaling. First, a quantitative analysis placed each family member from flies, mice, and nematodes into the Activin, BMP, or TGF-b subfamily. Second, we defined the prodomain and ligand via the consensus cleavage site. Third, we generated alignments and trees from the prodomain, ligand, and full-length sequences independently for each subfamily. Prodomain alignments revealed that six structural features of 17 are well conserved: three in the straitjacket and three in the arm. Alignments also revealed unexpected cysteine conservation in the “LTBP-Association region” upstream of the straitjacket and in b8 of the bowtie in 14 proteins from all three subfamilies. In prodomain trees, eight clusters across all three subfamilies were present that were not seen in the ligand or full- length trees, suggesting prodomain-mediated cross-subfamily heterodimerization. Consistency between cysteine conservation and prodomain clustering provides support for heterodimerization predictions. Overall, our analysis suggests that cross-subfamily interac- tions are more common than currently appreciated and our predictions generate numerous testable hypotheses about TGF-b function and evolution. KEYWORDS Activin; alignments/trees; arm/bowtie/straitjacket; BMP; cleavage site; heterodimer ECRETED TGF-b family members perform a myriad of to cell surface receptors. -
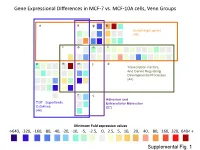
Supplementary Data
Gene Expressional Differences in MCF-7 vs. MCF-10A cells, Venn Groups e k g b Smad target genes (48) l o n i a h m j d Transcription Factors, And Genes Regulating Developmental Processes (44) f c Adhesion and TGF Superfamily Extracellular Molecules Cytokines (27) (44) Minimum Fold expression values ->640, -320, -160, -80, -40, -20, -10, -5, -2.5, 0, 2.5, 5, 10, 20, 40, 80, 160, 320, 640< + Supplemental Fig. 1 Supplemental Table 1. Change of Basal Gene Expressional values in MCF-7 as compared to MCF-10A cell line. Fold T-test Gene # GeneBank Symbol Up/Down p-Value Description /Position Regulation Venn Group a 01 /A01 NM_001105 ACVR1 -4.96 0.0000 Activin A receptor, type I 02 /A02 NM_001616 ACVR2A -2.62 0.0007 Activin A receptor, type IIA 03 /A03 NM_000020 ACVRL1 -1.25 0.7013 Activin A receptor type II-like 1 05 /A05 NM_020547 AMHR2 1.08 0.8043 Anti-Mullerian hormone receptor, type II 16 /B04 NM_004329 BMPR1A -2.40 0.0011 Bone morphogenetic protein receptor, type IA 17 /B05 NM_001203 BMPR1B -7.37 0.0000 Bone morphogenetic protein receptor, type IB 36 /C12 NM_000557 GDF5 -1.38 0.4911 Growth differentiation factor 5 (cartilage-derived morphogenetic protein-1) 37 /D01 NM_001001557 GDF6 1.12 0.9002 Growth differentiation factor 6 38 /D02 NM_182828 GDF7 -1.52 0.4995 Growth differentiation factor 7 53 /E05 NM_020997 LEFTY1 -1.76 0.0529 Left-right determination factor 1 59 /E11 NM_018055 NODAL -3.62 0.1290 Nodal homolog (mouse) 77 /G05 NM_003238 TGFB2 -4.52 0.0566 Transforming growth factor, beta 2 78 /G06 NM_003239 TGFB3 -1.12 0.2902 Transforming -

CER1 Is a Common Target of WNT and NODAL Signaling Pathways in Human Embryonic Stem Cells
795-799 24/3/06 13:04 Page 795 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 17: 795-799, 2006 795 CER1 is a common target of WNT and NODAL signaling pathways in human embryonic stem cells MASUKO KATOH1 and MASARU KATOH2 1M&M Medical BioInformatics, Hongo 113-0033; 2Genetics and Cell Biology Section, National Cancer Center Research Institute, Tokyo 104-0045, Japan Received January 3, 2006; Accepted February 7, 2006 Abstract. Nodal and BMP signaling pathways network with embryogenesis, underwent protein evolution as well as WNT signaling pathway during embryogenesis and carcino- promoter evolution. These facts indicate that molecular genesis. CER1 (Cerberus 1) and GREM3 (CKTSF1B3 or evolution of CER1 orthologs contributes to the significantly CER2) inhibit NODAL signaling through ACVR1B (ALK4) divergent scenarios of early embryogenesis in primates and or ACVR1C (ALK7) to SMAD2 or SMAD3. GREM1 rodents. (CKTSF1B1) inhibits BMP signaling through BMPR1A (ALK3), BMPR1B (ALK6) or ACVR1 (ALK2) to SMAD1, Introduction SMAD5 or SMAD8. CER1, GREM1 and GREM3 are DAN domain (DAND) family members; however, transcriptional TGFB1, TGFB2, TGFB3, NODAL, LEFTY1, LEFTY2, INHA, regulation of DAND family members by canonical WNT INHBA, INHBB, INHBC, INHBE, AMH, BMP2, BMP3, signaling pathway remains unclear. We searched for the BMP4, BMP5, BMP6, BMP7, BMP8A, BMP8B, BMP10, TCF/LEF-binding site within the promoter region of DAND BMP15, GDF1, GDF2, GDF3, GDF5, GDF6, GDF7, GDF8, family genes, including CER1, GREM1, GREM2, GREM3 and GDF9, GDF10, GDF11, and GDF15 are TGFß superfamily NBL1. Because triple TCF/LEF-binding sites were identified genes within the human genome (http://www.gene.ucl.ac.uk). within human CER1 promoter by using bioinformatics and TGFß signals are transduced through type I receptor TGFBR1 human intelligence, comparative genomics analyses on CER1 and type II receptor TGFBR2 to phosphorylate R-SMAD orthologs were further performed. -

Bone Morphogenetic Proteins in Vascular Homeostasis and Disease
Downloaded from http://cshperspectives.cshlp.org/ on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Bone Morphogenetic Proteins in Vascular Homeostasis and Disease Marie-Jose´Goumans,1 An Zwijsen,2,3 Peter ten Dijke,1,4 and Sabine Bailly5,6,7 1Department of Molecular Cell Biology, Leiden University Medical Center, 2300 RC Leiden, The Netherlands 2VIB Center for the Biology of Disease, 3000 Leuven, Belgium 3KU Leuven Department of Human Genetics, 3000 Leuven, Belgium 4Cancer Genomics Centre Netherlands, Leiden University Medical Center, 2300 RC Leiden, The Netherlands 5Institut National de la Sante´ et de la Recherche Me´cale (INSERM), U1036, 38000 Grenoble, France 6Laboratoire Biologie du Cancer et de l’Infection, Commissariat a` l’E´nergie Atomique et aux Energies Alternatives, Biosciences and Biotechnology Institute of Grenoble, 38000 Grenoble, France 7University of Grenoble Alpes, 38000 Grenoble, France Correspondence: [email protected]; [email protected] It is well established that control of vascular morphogenesis and homeostasis is regulated by vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), Delta-like 4 (Dll4), angiopoietin, and ephrin signaling. It has become clear that signaling by bone morphogenetic proteins (BMPs), which have a long history of studies in bone and early heart development, are also essential for regulating vascular function. Indeed, mutations that cause deregulated BMP signaling are linked to two human vascular diseases, hereditary hemorrhagic telangiectasia and pulmonary arterial hypertension. These observations are corroborated by data obtained with vascular cells in cell culture and in mouse models. BMPs are required for normal endothelial cell differentiation and for venous/arterial and lymphatic specification.