Visualizing Protein Structures - Tools and Trends Xavier Martinez, Matthieu Chavent, Marc Baaden
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Real-Time Pymol Visualization for Rosetta and Pyrosetta
Real-Time PyMOL Visualization for Rosetta and PyRosetta Evan H. Baugh1, Sergey Lyskov1, Brian D. Weitzner1, Jeffrey J. Gray1,2* 1 Department of Chemical and Biomolecular Engineering, The Johns Hopkins University, Baltimore, Maryland, United States of America, 2 Program in Molecular Biophysics, The Johns Hopkins University, Baltimore, Maryland, United States of America Abstract Computational structure prediction and design of proteins and protein-protein complexes have long been inaccessible to those not directly involved in the field. A key missing component has been the ability to visualize the progress of calculations to better understand them. Rosetta is one simulation suite that would benefit from a robust real-time visualization solution. Several tools exist for the sole purpose of visualizing biomolecules; one of the most popular tools, PyMOL (Schro¨dinger), is a powerful, highly extensible, user friendly, and attractive package. Integrating Rosetta and PyMOL directly has many technical and logistical obstacles inhibiting usage. To circumvent these issues, we developed a novel solution based on transmitting biomolecular structure and energy information via UDP sockets. Rosetta and PyMOL run as separate processes, thereby avoiding many technical obstacles while visualizing information on-demand in real-time. When Rosetta detects changes in the structure of a protein, new coordinates are sent over a UDP network socket to a PyMOL instance running a UDP socket listener. PyMOL then interprets and displays the molecule. This implementation also allows remote execution of Rosetta. When combined with PyRosetta, this visualization solution provides an interactive environment for protein structure prediction and design. Citation: Baugh EH, Lyskov S, Weitzner BD, Gray JJ (2011) Real-Time PyMOL Visualization for Rosetta and PyRosetta. -

A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Mohebifar and Sajadi J Cheminform (2015) 7:56 DOI 10.1186/s13321-015-0101-7 SOFTWARE Open Access Chemozart: a web‑based 3D molecular structure editor and visualizer platform Mohamad Mohebifar* and Fatemehsadat Sajadi Abstract Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimen- sional structures of molecules. Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user- friendly interface is composed. More than 30 packages are used to compose this application which adds enough flex- ibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context. Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (applica- tion programming interface). JavaScript libraries which allow creation of web pages containing interactive three- dimensional molecular structures has also been made available. -

64-194 Projekt Parallelrechnerevaluation Abschlussbericht
64-194 Projekt Parallelrechnerevaluation Abschlussbericht Automatisierte Build-Tests für Spack Sarah Lubitz Malte Fock [email protected] [email protected] Studiengang: LA Berufliche Schulen – Informatik Studiengang LaGym - Informatik Matr.-Nr. 6570465 Matr.-Nr. 6311966 Inhaltsverzeichnis i Inhaltsverzeichnis 1 Einleitung1 1.1 Motivation.....................................1 1.2 Vorbereitungen..................................2 1.2.1 Spack....................................2 1.2.2 Python...................................2 1.2.3 Unix-Shell.................................3 1.2.4 Slurm....................................3 1.2.5 GitHub...................................4 2 Code 7 2.1 Automatisiertes Abrufen und Installieren von Spack-Paketen.......7 2.1.1 Python Skript: install_all_packets.py..................7 2.2 Batch-Jobskripte..................................9 2.2.1 Test mit drei Paketen........................... 10 2.3 Analyseskript................................... 11 2.3.1 Test mit 500 Paketen........................... 12 2.3.2 Code zur Analyse der Error-Logs, forts................. 13 2.4 Hintereinanderausführung der Installationen................. 17 2.4.1 Wrapper Skript.............................. 19 2.5 E-Mail Benachrichtigungen........................... 22 3 Testlauf 23 3.1 Durchführung und Ergebnisse......................... 23 3.2 Auswertung und Schlussfolgerungen..................... 23 3.3 Fazit und Ausblick................................ 24 4 Schulische Relevanz 27 Literaturverzeichnis -

Computational Modelling of Wet Adhesive Mussel Foot Proteins (Bivalvia): Insights Into the Evolutionary Convolution in Diverse Perspectives P
www.nature.com/scientificreports OPEN Computational modelling of wet adhesive mussel foot proteins (Bivalvia): Insights into the evolutionary convolution in diverse perspectives P. P. Anand & Y. Shibu Vardhanan Underwater adhesion in mussels (Bivalvia) is an extreme adaptation to achieve robust and frm wet adhesion in the freshwater/brackish/ocean, which biochemically shaped through millions of years. The protein-based adhesion has huge prospective in various felds like industry, medical, etc. Currently, no comprehensive records related to the systematic documentation of structural and functional properties of Mussel foot proteins (Mfps). In this study, we identifed the nine species of bivalves in which the complete sequence of at least one adhesive protein is known. The insilico characterization revealed the specifc physio-chemical structural and functional characters of each Mfps. The evolutionary analyses of selected bivalves are mainly based on Mfps, Mitogenome, and TimeTree. The outcome of the works has great applications for designing biomimetic materials in future. Some groups of mussels are capable to produce proteinaceous glue- like sticky material known as byssus thread made by an array of foot proteins (fps). Tis byssus contains mainly four parts i.e. Plaque, thread, stem, and root. Individual threads proximally merged together to form stem and base of the stem (root) deeply anchored at the base of animal foot. Each byssus threads terminating distally with a fattened plaque which mediates adhesion to the substratum1–4. Each part of the byssus thread complex formed by the auto-assembly of secretory products originating from four distinct glands enclosed in the mussel foot4,5. Tese mussel foot protein (Mfps), mastered the ability to binding the diverse substratum by using adhesive plaques. -

Water and Salt at the Lipid-Solvent Interface
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School April 2019 Water and Salt at the Lipid-Solvent Interface James M. Kruczek University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Physics Commons Scholar Commons Citation Kruczek, James M., "Water and Salt at the Lipid-Solvent Interface" (2019). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/8380 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Water and Salt at the Lipid-Solvent Interface by James M. Kruczek A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Department of Physics College of Arts and Sciences University of South Florida Major Professor: Sagar A. Pandit, Ph.D. Ullah, Ghanim, Ph.D. Robert S. Hoy, Ph.D. Jianjun Pan, Ph.D. Yicheng Tu, Ph.D. Date of Approval: March 26, 2019 Keywords: Lipid Bilayer, Ionic Solvents, Ether Lipids, Molecular Simulations Copyright ⃝c 2018, James M. Kruczek Dedication To my wife Nicole, without whom none of this would be possible. To my father Michael, who labored for his family till his last days. To my family, for all of their support. Acknowledgments The work presented in this document would not be possible without the assistance of many academic professionals. In particular, I would like to acknowledge my major advisor Dr. -

How to Print a Crystal Structure Model in 3D Teng-Hao Chen,*A Semin Lee, B Amar H
Electronic Supplementary Material (ESI) for CrystEngComm. This journal is © The Royal Society of Chemistry 2014 How to Print a Crystal Structure Model in 3D Teng-Hao Chen,*a Semin Lee, b Amar H. Flood b and Ognjen Š. Miljanić a a Department of Chemistry, University of Houston, 112 Fleming Building, Houston, Texas 77204-5003, United States b Department of Chemistry, Indiana University, 800 E. Kirkwood Avenue, Bloomington, Indiana 47405, United States Supporting Information Instructions for Printing a 3D Model of an Interlocked Molecule: Cyanostars .................................... S2 Instructions for Printing a "Molecular Spinning Top" in 3D: Triazolophanes ..................................... S3 References ....................................................................................................................................................................... S4 S1 Instructions for Printing a 3D Model of an Interlocked Molecule: Cyanostars Mechanically interlocked molecules such as rotaxanes and catenanes can also be easily printed in 3D. As long as the molecule is correctly designed and modeled, there is no need to assemble and glue the components after printing—they are printed inherently as mechanically interlocked molecules. We present the preparation of a cyanostar- based [3]rotaxane as an example. A pair of sandwiched cyanostars S1 (CS ) derived from the crystal structure was "cleaned up" in the molecular computation software Spartan ʹ10, i.e. disordered atoms were removed. A linear molecule was created and threaded through the cavity of two cyanostars (Figure S1a). Using a space filling view of the molecule, the three components were spaced sufficiently far apart to ensure that they did not make direct contact Figure S1 . ( a) Side and top view of two cyanostars ( CS ) with each other when they are printed. threaded with a linear molecule. ( b) Side and top view of a Bulky stoppers were added to each [3]rotaxane. -

Conformational and Chemical Selection by a Trans-Acting Editing
Correction BIOCHEMISTRY Correction for “Conformational and chemical selection by a trans- acting editing domain,” by Eric M. Danhart, Marina Bakhtina, William A. Cantara, Alexandra B. Kuzmishin, Xiao Ma, Brianne L. Sanford, Marija Kosutic, Yuki Goto, Hiroaki Suga, Kotaro Nakanishi, Ronald Micura, Mark P. Foster, and Karin Musier-Forsyth, which was first published August 2, 2017; 10.1073/pnas.1703925114 (Proc Natl Acad Sci USA 114:E6774–E6783). The authors note that Oscar Vargas-Rodriguez should be added to the author list between Brianne L. Sanford and Marija Kosutic. Oscar Vargas-Rodriguez should be credited with de- signing research, performing research, and analyzing data. The corrected author line, affiliation line, and author contributions appear below. The online version has been corrected. The authors note that the following statement should be added to the Acknowledgments: “We thank Daniel McGowan for assistance with preparing mutant proteins.” Eric M. Danharta,b, Marina Bakhtinaa,b, William A. Cantaraa,b, Alexandra B. Kuzmishina,b,XiaoMaa,b, Brianne L. Sanforda,b, Oscar Vargas-Rodrigueza,b, Marija Kosuticc,d,YukiGotoe, Hiroaki Sugae, Kotaro Nakanishia,b, Ronald Micurac,d, Mark P. Fostera,b, and Karin Musier-Forsytha,b aDepartment of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210; bCenter for RNA Biology, The Ohio State University, Columbus, OH 43210; cInstitute of Organic Chemistry, Leopold Franzens University, A-6020 Innsbruck, Austria; dCenter for Molecular Biosciences, Leopold Franzens University, A-6020 Innsbruck, Austria; and eDepartment of Chemistry, Graduate School of Science, The University of Tokyo, Tokyo 113-0033, Japan Author contributions: E.M.D., M.B., W.A.C., B.L.S., O.V.-R., M.P.F., and K.M.-F. -

Tracking of Bacterial Metabolism with Azidated Precursors and Click- Chemistry”
Tracking of bacterial metabolism with azidated precursors and click-chemistry Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften vorgelegt beim Fachbereich für Biowissenschaften (15) der Johann Wolfang Goethe Universität in Frankfurt am Main von Alexander J. Pérez aus Nürnberg Frankfurt am Main 2015 Dekanin: Prof. Dr. Meike Piepenbring Gutachter: Prof. Dr. Helge B. Bode Zweitgutachter: Prof. Dr. Joachim W. Engels Datum der Disputation: 2 Danksagung Ich danke meinen Eltern für die stete und vielseitige Unterstützung, deren Umfang ich sehr zu schätzen weiß. Herrn Professor Dr. Helge B. Bode gilt mein besonderer Dank für die Übernahme als Doktorand und für die Gelegenheit meinen Horizont in diesen mich stets faszinierenden Themenbereich in dieser Tiefe erweitern zu lassen. Seine persönliche und fachliche Unterstützung bei der Projektwahl und der entsprechenden Umsetzung ist in dieser Form eine Seltenheit und ich bin mir dieser Tatsache voll bewusst. Gerade die zusätzlich erworbenen Kenntnisse im Bereich der Biologie, sowie der Wert interdisziplinärer Zusammenarbeit ist mir durch zahlreiche freundliche und wertvolle Mitglieder der Arbeitsgruppe bewusst geworden und viele zündenden Ideen wären ohne sie womöglich nie aufgekommen. Einen besonderen Dank möchte ich in diesem Kontext Wolfram Lorenzen und Sebastian Fuchs, die gerade in der Anfangszeit eine große Hilfe waren, ausdrücken. Dies gilt ebenso für die „N100-Crew“ und sämtliche Freunde, die in dieser Zeit zu mir standen und diesen Lebensabschnitt unvergesslich gemacht haben. -
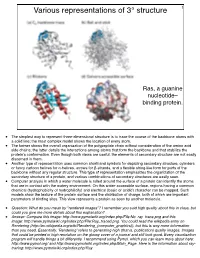
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

Package Name Software Description Project
A S T 1 Package Name Software Description Project URL 2 Autoconf An extensible package of M4 macros that produce shell scripts to automatically configure software source code packages https://www.gnu.org/software/autoconf/ 3 Automake www.gnu.org/software/automake 4 Libtool www.gnu.org/software/libtool 5 bamtools BamTools: a C++ API for reading/writing BAM files. https://github.com/pezmaster31/bamtools 6 Biopython (Python module) Biopython is a set of freely available tools for biological computation written in Python by an international team of developers www.biopython.org/ 7 blas The BLAS (Basic Linear Algebra Subprograms) are routines that provide standard building blocks for performing basic vector and matrix operations. http://www.netlib.org/blas/ 8 boost Boost provides free peer-reviewed portable C++ source libraries. http://www.boost.org 9 CMake Cross-platform, open-source build system. CMake is a family of tools designed to build, test and package software http://www.cmake.org/ 10 Cython (Python module) The Cython compiler for writing C extensions for the Python language https://www.python.org/ 11 Doxygen http://www.doxygen.org/ FFmpeg is the leading multimedia framework, able to decode, encode, transcode, mux, demux, stream, filter and play pretty much anything that humans and machines have created. It supports the most obscure ancient formats up to the cutting edge. No matter if they were designed by some standards 12 ffmpeg committee, the community or a corporation. https://www.ffmpeg.org FFTW is a C subroutine library for computing the discrete Fourier transform (DFT) in one or more dimensions, of arbitrary input size, and of both real and 13 fftw complex data (as well as of even/odd data, i.e. -

Visualizing Protein Structures-Tools and Trends
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 January 2020 Visualizing protein structures - tools and trends 1,2 3 1,2 X. Martinez , M. Chavent , M. Baaden 1) CNRS, Université de Paris, UPR 9080, Laboratoire de Biochimie Théorique, 13 rue Pierre et Marie Curie, F-75005, Paris, France 2) Institut de Biologie Physico-Chimique-Fondation Edmond de Rotschild, PSL Research University, Paris, France 3) Institut de Pharmacologie et de Biologie Structurale IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France Abstract Molecular visualisation is fundamental in the current scientific literature, textbooks and dissemination materials, forming an essential support for presenting results, reasoning on and formulating hypotheses related to molecular structure. Visual exploration has become easily accessible on a broad variety of platforms thanks to advanced software tools that render a great service to the scientific community. These tools are often developed across disciplines bridging computer science, biology and chemistry. Here we first describe a few Swiss Army knives geared towards protein visualisation for everyday use with an existing large user base, then focus on more specialised tools for peculiar needs that are not yet as broadly known. Our selection is by no means exhaustive, but reflects a diverse snapshot of scenarios that we consider informative for the reader. We end with an account of future trends and perspectives. Keywords Molecular Graphics, Protein visualization, Software tools, Virtual reality Introduction Many parts of science rely on a visualization-driven cycle of experimentation, reasoning, conjecture and validation, even more so in relation with structural biology and biophysics. Molecular visualization (1) in particular is now broadly used in many contexts, with the purpose of illustration in the scientific literature or the aim to gain insight about primary research data. -

Engine for Molecule Visualization in a Web Browser
MASARYKOVA UNIVERZITA FAKULTA}w¡¢£¤¥¦§¨ INFORMATIKY !"#$%&'()+,-./012345<yA| Engine for Molecule Visualization in a Web Browser MASTER’S THESIS Jaromír Svoboda Brno, spring 2014 Declaration Hereby I declare, that this paper is my original authorial work, which I have worked out by my own. All sources, references and literature used or excerpted during elaboration of this work are properly cited and listed in complete reference to the due source. Jaromír Svoboda Advisor: RNDr. David Sehnal ii Acknowledgement I would like to thank my supervisor RNDr. David Sehnal for patient guidance and informed advice throughout writing this thesis. iii Abstract The main focus of this master’s thesis is the design and implementa- tion of lightweight molecular visualization engine (called LiveMol) in form of a JavaScript library. The engine utilizes widely adopted WebGL API to display GPU-accelerated graphics in web browsers. Due to the size of complex protein molecules, the primary goal is high performance. Furthermore, the design of LiveMol enables users to extend the core functionality by simply defining custom coloring schemes of molecule models or implement completely new visual- ization modes. iv Keywords molecular visualization, protein, secondary structure, WebGL, LiveMol v Contents I INTRODUCTION 1 1 Introduction ............................2 II CURRENT DEVELOPMENTS 3 2 Current Molecular Visualization Software ..........4 2.1 Desktop Applications ....................4 2.1.1 Visual Molecular Dynamic . .4 2.1.2 PyMOL . .4 2.1.3 RasMol/OpenRasMol . .5 2.1.4 BALL/BALLView . .5 2.1.5 Gabedit . .5 2.1.6 QuteMol . .5 2.1.7 Avogadro . .6 2.2 JavaScript-based Web Applications ............6 2.2.1 Jmol/JSmol .