22S:152 Applied Linear Regression Chapter 5: Ordinary Least Squares
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
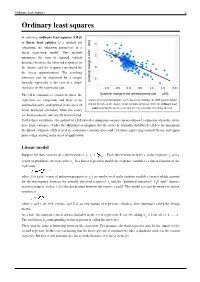
Ordinary Least Squares 1 Ordinary Least Squares
Ordinary least squares 1 Ordinary least squares In statistics, ordinary least squares (OLS) or linear least squares is a method for estimating the unknown parameters in a linear regression model. This method minimizes the sum of squared vertical distances between the observed responses in the dataset and the responses predicted by the linear approximation. The resulting estimator can be expressed by a simple formula, especially in the case of a single regressor on the right-hand side. The OLS estimator is consistent when the regressors are exogenous and there is no Okun's law in macroeconomics states that in an economy the GDP growth should multicollinearity, and optimal in the class of depend linearly on the changes in the unemployment rate. Here the ordinary least squares method is used to construct the regression line describing this law. linear unbiased estimators when the errors are homoscedastic and serially uncorrelated. Under these conditions, the method of OLS provides minimum-variance mean-unbiased estimation when the errors have finite variances. Under the additional assumption that the errors be normally distributed, OLS is the maximum likelihood estimator. OLS is used in economics (econometrics) and electrical engineering (control theory and signal processing), among many areas of application. Linear model Suppose the data consists of n observations { y , x } . Each observation includes a scalar response y and a i i i vector of predictors (or regressors) x . In a linear regression model the response variable is a linear function of the i regressors: where β is a p×1 vector of unknown parameters; ε 's are unobserved scalar random variables (errors) which account i for the discrepancy between the actually observed responses y and the "predicted outcomes" x′ β; and ′ denotes i i matrix transpose, so that x′ β is the dot product between the vectors x and β. -

Linear Regression
DATA MINING 2 Linear and Logistic Regression Riccardo Guidotti a.a. 2019/2020 Regression • Given a dataset containing N observations Xi, Yi i = 1, 2, …, N • Regression is the task of learning a target function f that maps each input attribute set X into a output Y. • The goal is to find the target function that can fit the input data with minimum error. • The error function can be expressed as • Absolute Error = ∑! |�! − �(�!)| " • Squared Error = ∑!(�! − � �! ) residuals Linear Regression Y • Linear regression is a linear approach to modeling the relationship between a dependent variable Y and one or more independent (explanatory) variables X. • The case of one explanatory variable is called simple linear regression. • For more than one explanatory variable, the process is called multiple linear X regression. • For multiple correlated dependent variables, the process is called multivariate linear regression. What does it mean to predict Y? • Look at X = 5. There are many different Y values at X=5. • When we say predict Y at X =5, we are really asking: • What is the expected value (average) of Y at X = 5? What does it mean to predict Y? • Formally, the regression function is given by E(Y|X=x). This is the expected value of Y at X=x. • The ideal or optimal predictor of Y based on X is thus • f(X) = E(Y | X=x) Simple Linear Regression Dependent Independent Variable Variable Linear Model: Y = mX + b � = �!� + �" Slope Intercept (bias) • In general, such a relationship may not hold exactly for the largely unobserved population • We call the unobserved deviations from Y the errors. -

Linear Regression
eesc BC 3017 statistics notes 1 LINEAR REGRESSION Systematic var iation in the true value Up to now, wehav e been thinking about measurement as sampling of values from an ensemble of all possible outcomes in order to estimate the true value (which would, according to our previous discussion, be well approximated by the mean of a very large sample). Givenasample of outcomes, we have sometimes checked the hypothesis that it is a random sample from some ensemble of outcomes, by plotting the data points against some other variable, such as ordinal position. Under the hypothesis of random selection, no clear trend should appear.Howev er, the contrary case, where one finds a clear trend, is very important. Aclear trend can be a discovery,rather than a nuisance! Whether it is adiscovery or a nuisance (or both) depends on what one finds out about the reasons underlying the trend. In either case one must be prepared to deal with trends in analyzing data. Figure 2.1 (a) shows a plot of (hypothetical) data in which there is a very clear trend. The yaxis scales concentration of coliform bacteria sampled from rivers in various regions (units are colonies per liter). The x axis is a hypothetical indexofregional urbanization, ranging from 1 to 10. The hypothetical data consist of 6 different measurements at each levelofurbanization. The mean of each set of 6 measurements givesarough estimate of the true value for coliform bacteria concentration for rivers in a region with that urbanization level. The jagged dark line drawn on the graph connects these estimates of true value and makes the trend quite clear: more extensive urbanization is associated with higher true values of bacteria concentration. -

Simple Linear Regression
The simple linear model Represents the dependent variable, yi, as a linear function of one Regression Analysis: Basic Concepts independent variable, xi, subject to a random “disturbance” or “error”, ui. yi β0 β1xi ui = + + Allin Cottrell The error term ui is assumed to have a mean value of zero, a constant variance, and to be uncorrelated with its own past values (i.e., it is “white noise”). The task of estimation is to determine regression coefficients βˆ0 and βˆ1, estimates of the unknown parameters β0 and β1 respectively. The estimated equation will have the form yˆi βˆ0 βˆ1x = + 1 OLS Picturing the residuals The basic technique for determining the coefficients βˆ0 and βˆ1 is Ordinary Least Squares (OLS). Values for the coefficients are chosen to minimize the sum of the ˆ ˆ squared estimated errors or residual sum of squares (SSR). The β0 β1x + estimated error associated with each pair of data-values (xi, yi) is yi uˆ defined as i uˆi yi yˆi yi βˆ0 βˆ1xi yˆi = − = − − We use a different symbol for this estimated error (uˆi) as opposed to the “true” disturbance or error term, (ui). These two coincide only if βˆ0 and βˆ1 happen to be exact estimates of the regression parameters α and β. The estimated errors are also known as residuals . The SSR may be written as xi 2 2 2 SSR uˆ (yi yˆi) (yi βˆ0 βˆ1xi) = i = − = − − Σ Σ Σ The residual, uˆi, is the vertical distance between the actual value of the dependent variable, yi, and the fitted value, yˆi βˆ0 βˆ1xi. -

Canada 2008 © 2008 Laura Duncan Library and Bibliotheque Et 1*1 Archives Canada Archives Canada Published Heritage Direction Du Branch Patrimoine De I'edition
EFFECT OF BLOCK FACE SHELL GEOMETRY AND GROUTING ON THE COMPRESSIVE STRENGTH OF CONCRETE BLOCK MASONRY by Laura J. Duncan A Thesis Submitted to the Faculty of Graduate Studies through Civil and Environmental Engineering in Partial Fulfillment of the Requirements for the Degree of Master of Applied Science at the University of Windsor Windsor, Ontario, Canada 2008 © 2008 Laura Duncan Library and Bibliotheque et 1*1 Archives Canada Archives Canada Published Heritage Direction du Branch Patrimoine de I'edition 395 Wellington Street 395, rue Wellington Ottawa ON K1A0N4 Ottawa ON K1A0N4 Canada Canada Your file Votre reference ISBN: 978-0-494-47089-3 Our file Notre reference ISBN: 978-0-494-47089-3 NOTICE: AVIS: The author has granted a non L'auteur a accorde une licence non exclusive exclusive license allowing Library permettant a la Bibliotheque et Archives and Archives Canada to reproduce, Canada de reproduire, publier, archiver, publish, archive, preserve, conserve, sauvegarder, conserver, transmettre au public communicate to the public by par telecommunication ou par Plntemet, prefer, telecommunication or on the Internet, distribuer et vendre des theses partout dans loan, distribute and sell theses le monde, a des fins commerciales ou autres, worldwide, for commercial or non sur support microforme, papier, electronique commercial purposes, in microform, et/ou autres formats. paper, electronic and/or any other formats. The author retains copyright L'auteur conserve la propriete du droit d'auteur ownership and moral rights in et des droits moraux qui protege cette these. this thesis. Neither the thesis Ni la these ni des extraits substantiels de nor substantial extracts from it celle-ci ne doivent etre imprimes ou autrement may be printed or otherwise reproduits sans son autorisation. -
On the Determination of Proper Regularization Parameter
Universität Stuttgart Geodätisches Institut On the determination of proper regularization parameter α-weighted BLE via A-optimal design and its comparison with the results derived by numerical methods and ridge regression Bachelorarbeit im Studiengang Geodäsie und Geoinformatik an der Universität Stuttgart Kun Qian Stuttgart, Februar 2017 Betreuer: Dr.-Ing. Jianqing Cai Universität Stuttgart Prof. Dr.-Ing. Nico Sneeuw Universität Stuttgart Erklärung der Urheberschaft Ich erkläre hiermit an Eides statt, dass ich die vorliegende Arbeit ohne Hilfe Dritter und ohne Benutzung anderer als der angegebenen Hilfsmittel angefertigt habe; die aus fremden Quellen direkt oder indirekt übernommenen Gedanken sind als solche kenntlich gemacht. Die Arbeit wurde bisher in gleicher oder ähnlicher Form in keiner anderen Prüfungsbehörde vorgelegt und auch noch nicht veröffentlicht. Ort, Datum Unterschrift III Abstract In this thesis, several numerical regularization methods and the ridge regression, which can help improve improper conditions and solve ill-posed problems, are reviewed. The deter- mination of the optimal regularization parameter via A-optimal design, the optimal uniform Tikhonov-Phillips regularization (a-weighted biased linear estimation), which minimizes the trace of the mean square error matrix MSE(ˆx), is also introduced. Moreover, the comparison of the results derived by A-optimal design and results derived by numerical heuristic methods, such as L-curve, Generalized Cross Validation and the method of dichotomy is demonstrated. According to the comparison, the A-optimal design regularization parameter has been shown to have minimum trace of MSE(ˆx) and its determination has better efficiency. VII Contents List of Figures IX List of Tables XI 1 Introduction 1 2 Least Squares Method and Ill-Posed Problems 3 2.1 The Special Gauss-Markov medel . -

Akaike's Information Criterion
ESCI 340 Biostatistical Analysis Model Selection with Information Theory "Far better an approximate answer to the right question, which is often vague, than an exact answer to the wrong question, which can always be made precise." − John W. Tukey, (1962), "The future of data analysis." Annals of Mathematical Statistics 33, 1-67. 1 Problems with Statistical Hypothesis Testing 1.1 Indirect approach: − effort to reject null hypothesis (H0) believed to be false a priori (statistical hypotheses are not the same as scientific hypotheses) 1.2 Cannot accommodate multiple hypotheses (e.g., Chamberlin 1890) 1.3 Significance level (α) is arbitrary − will obtain "significant" result if n large enough 1.4 Tendency to focus on P-values rather than magnitude of effects 2 Practical Alternative: Direct Evaluation of Multiple Hypotheses 2.1 General Approach: 2.1.1 Develop multiple hypotheses to answer research question. 2.1.2 Translate each hypothesis into a model. 2.1.3 Fit each model to the data (using least squares, maximum likelihood, etc.). (fitting model ≅ estimating parameters) 2.1.4 Evaluate each model using information criterion (e.g., AIC). 2.1.5 Select model that performs best, and determine its likelihood. 2.2 Model Selection Criterion 2.2.1 Akaike Information Criterion (AIC): relates information theory to maximum likelihood ˆ AIC = −2loge[L(θ | data)]+ 2K θˆ = estimated model parameters ˆ loge[L(θ | data)] = log-likelihood, maximized over all θ K = number of parameters in model Select model that minimizes AIC. 2.2.2 Modification for complex models (K large relative to sample size, n): 2K(K +1) AIC = −2log [L(θˆ | data)]+ 2K + c e n − K −1 use AICc when n/K < 40. -
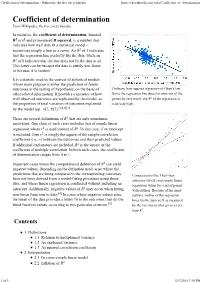
Coefficient of Determination - Wikipedia, the Free Encyclopedia
Coefficient of determination - Wikipedia, the free encyclopedia https://en.wikipedia.org/wiki/Coefficient_of_determination From Wikipedia, the free encyclopedia In statistics, the coefficient of determination, denoted R2 or r2 and pronounced R squared, is a number that indicates how well data fit a statistical model – sometimes simply a line or a curve. An R2 of 1 indicates that the regression line perfectly fits the data, while an R2 of 0 indicates that the line does not fit the data at all. This latter can be because the data is utterly non-linear, or because it is random. It is a statistic used in the context of statistical models whose main purpose is either the prediction of future outcomes or the testing of hypotheses, on the basis of Ordinary least squares regression of Okun's law. other related information. It provides a measure of how Since the regression line does not miss any of the well observed outcomes are replicated by the model, as points by very much, the R2 of the regression is the proportion of total variation of outcomes explained relatively high. by the model (pp. 187, 287).[1][2][3] There are several definitions of R2 that are only sometimes equivalent. One class of such cases includes that of simple linear regression where r2 is used instead of R2. In this case, if an intercept is included, then r2 is simply the square of the sample correlation coefficient (i.e., r) between the outcomes and their predicted values. If additional explanators are included, R2 is the square of the coefficient of multiple correlation. -

Statistical Modeling of Fracture Toughness Data
Statistical Modeling Of Fracture Toughness Data by Guru Prakash A thesis presented to the University of Waterloo in fulfillment of the thesis requirement for the degree of Master of Applied Science in Civil and Environmental Engineering Waterloo, Ontario, Canada, 2007 © Guru Prakash, 2007 AUTHOR'S DECLARATION I hereby declare that I am the sole author of this thesis. This is a true copy of the thesis, including any required final revisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Abstract The fracture toughness of the zirconium alloy (Zr-2.5Nb) is an important parameter in determining the flaw tolerance for operation of pressure tubes in reactor. Fracture toughness data have been generated by performing rising pressure burst tests on sections of pressure tubes removed from operating reactors. The test data were used to generate a lower-bound fracture toughness curve, which is used in defining the operational limits of pressure tubes. The thesis presents a comprehensive statistical analysis of burst test data and develops a multivariate statistical model to relate toughness with material chemistry, mechanical properties, and operational history. The proposed model can be useful in predicting fracture toughness of specific in-service pressure tubes, thereby minimizing conservatism associated with a generic lower-bound approach. iii Acknowledgements I would like to thank my supervisor Professor Mahesh D. Pandey for his support, encouragement, guidance, and patience during my study at the University of Waterloo. I would like to thank Professor Galena Morozova for providing me Teaching Assistantship for the CIVE153 course. -

Note 17: F-Tests of Linear Coefficient Restrictions … Page 1 of 33 Pages ECONOMICS 351* -- NOTE 17 M.G
ECONOMICS 351* -- NOTE 17 M.G. Abbott ECON 351* -- NOTE 17 F-Tests of Linear Coefficient Restrictions: A General Approach 1. Introduction • The population regression equation (PRE) for the general multiple linear regression model takes the form: Yi= β 0 + β 1XX 1i + β 2 2i + L + βkX ki+ u i (1) where ui is an iid (independently and identically distributed) random error term. PRE (1) constitutes the unrestricted model. • The OLS sample regression equation (OLS-SRE) for equation (1) can be written as ˆ ˆ ˆ ˆ ˆ Yi=β+β 0 1X 1i +β 2X 2i +L +βkX ki +uˆ i =Y i + uˆ i (i = 1, ..., N) (2) where (1) the OLS estimated (or predicted) values of Yi , or the OLS sample regression function (OLS-SRF), are ˆ ˆ ˆ ˆ ˆ Yi= β 0 + β 1X 1i + β 2X 2i +L + βk)NX ki , . ( , 1 = i (2) the OLS residuals are ˆ ˆ ˆ ˆ ˆ uˆ i= Y i −YY i = i −β−β 0 11iX −β 22iX −L −βkX ki (i = 1, ..., N) ˆ (3) the βj are the OLS estimators of the corresponding population regression coefficients βj (j = 0, 1, ..., k). ECON 351* -- Note 17: F-Tests of Linear Coefficient Restrictions … Page 1 of 33 pages ECONOMICS 351* -- NOTE 17 M.G. Abbott ˆ ˆ ˆ ˆ ˆ Yi=β+β 0 1X 1i +β 2X 2i +L +βkX ki +uˆ i =Y i + uˆ i (i = 1, ..., N) ( ) 2 • The OLS decomposition equation for the OLS-SRE (2) is N N N N N 2 ˆ ˆ ˆ 2 ∑yi = β1 ∑x 1i y i + β2 ∑ x 2i y i +L + βk∑x ki y i + ∑ uˆ i i= 1 i= 1 i= 1 i= 1 i= 1 ⏐ ↑______________________________↑ ⏐ ⏐ ⏐ ⏐ ↓ ↓ ↓ TSS = ESS1 + RSS1 (N − ) 1 K( − )1 N(−K) where the figures in parentheses are the respective degrees of freedom for the Total Sum of Squares (TSS), the Explained (Regression) Sum of Squares (ESS1) and the Residual (Error) Sum of Squares (RSS1). -

Package 'Cusp'
Package ‘cusp’ August 10, 2015 Type Package Title Cusp-Catastrophe Model Fitting Using Maximum Likelihood Version 2.3.3 Imports stats, graphics, grDevices, utils Author Raoul P. P. P. Grasman [aut, cre, cph] Maintainer Raoul Grasman <[email protected]> LazyData yes NeedsCompilation yes Suggests plot3D Description Cobb's maximum likelihood method for cusp-catastrophe modeling (Grasman, van der Maas, & Wagenmakers, 2009, JSS, 32:8; Cobb, L, 1981, Behavioral Science, 26:1, 75--78). Includes a cusp() function for model fitting, and several utility functions for plotting, and for comparing the model to linear regression and logistic curve models. License GPL-2 Repository CRAN Repository/R-Forge/Project cusp Repository/R-Forge/Revision 21 Repository/R-Forge/DateTimeStamp 2015-07-29 11:18:21 Date/Publication 2015-08-10 18:31:48 R topics documented: cusp-package . .2 attitudes . .4 cusp.............................................5 cusp.bifset . .8 cusp.extrema . .9 cusp.logist . 10 cusp.nc . 12 1 2 cusp-package cusp.nlogLike . 13 cusp3d . 15 cusp3d.surface . 16 dcusp . 19 draw.cusp.bifset . 20 oliva............................................. 22 plot.cusp . 23 plotCuspBifurcation . 24 plotCuspDensities . 26 plotCuspResidfitted . 27 predict.cusp . 27 summary.cusp . 29 vcov.cusp . 32 zeeman . 33 Index 35 cusp-package Cusp Catastrophe Modeling Description Fits cusp catastrophe to data using Cobb’s maximum likelihood method with a different algorithm. The package contains utility functions for plotting, and for comparing the model to linear regression and logistic curve models. The package allows for multivariate response subspace modelling in the sense of the GEMCAT software of Oliva et al. Details Package: cusp Type: Package Version: 2.0 Date: 2008-02-14 License: GNU GPL v2 (or higher) This package helps fitting Cusp catastrophy models to data, as advanced in Cobb et al. -

2019 Akaike Information Criterion in Weighted Regression of Immittance
Accepted version of Electrochim. Acta, 317 (2019) 648-653. doi: 10.1016/j.electacta.2019.06.030 © 2019. This manuscript version is made available under the CC-BY-NC-ND 4.0 license http://creativecommons.org/licences/by-nc-nd/4.0/ The Akaike information criterion in weighted regression of immittance data a a ,b Mats Ingdal ,RoyJohnsen, David A. Harrington∗ a Department of Mechanical and Industrial Engineering, Norwegian University of Science and Technology, Trondheim, 7491, Norway. b Department of Chemistry, University of Victoria, Victoria, British Columbia, V8W 2Y2, Canada. Abstract The Akaike Information Criterion (AIC) is a powerful way to distinguish be- tween models. It considers both the goodness-of-fit and the number of parame- ters in the model, but has been little used for immittance data. Application in the case of weighted complex nonlinear least squares regression, as typically used in fitting impedance or admittance data, is considered here. AIC can be used to compare different weighting schemes as well as different models. These ideas are tested for simulated and real transadmittance data for hydrogen diffusion through an iron foil in a Devanathan-Stachurski cell. Key words: Akaike Information Criterion, weighted regression, error structure, impedance, maximum likelihood Introduction The Akaike Information Criterion (AIC) was developed by Akaike in 1973 [1] and is widely used in modeling of biological systems to decide between different models [2]. For models with the same number of adjustable parameters, it selects the model with the best least-squares fit. Models with more parameters typically fit better, but addition of more parameters may not be statistically justifiable.