Druggable Transient Pockets in Protein Kinases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Allosteric Regulation in Drug Design
Mini Review Curr Trends Biomedical Eng & Biosci Volume 4 Issue 1 - May 2017 Copyright © All rights are reserved by Ashfaq Ur Rehman DOI: 10.19080/CTBEB.2017.04.5555630 Allosteric regulation in drug design Ashfaq Ur Rehman1,2*, Shah Saud3, Nasir Ahmad4, Abdul Wadood2 and R Hamid5 1State Key Laboratory of Microbial Metabolism, Department of Bioinformatics and Biostatistics, China 2Department of Biochemistry, Abdul Wali Khan University Mardan, Pakistan 3Laboratory of Analytical Biochemistry and Bio separation, Shanghai Jiao Tong University, China 4Department of Chemistry, Islama College University Peshawar, Pakistan 5Department of Bioinformatics, Muhammad Ali Jinnah University Islamabad, Pakistan Submission: May 02, 2017; Published: May 23, 2017 *Corresponding author: Ashfaq Ur Rehman, State Key Laboratory of Microbial Metabolism, Department of Bioinformatics and Biostatistics, Shanghai Jiao Tong University, 800 Dongchuan Road, Shanghai 200240, China, Tel: ; Fax: 86-21-34204348; Email: Abstract mechanism, which are initiated through attachment of ligand or inhibitors with the protein or enzymes other than active (orthosteric) sites. ThisProtein mini review and enzymes involved play mechanism, significant types roles and in importancebiological processes of allosteric of all regulations living organisms; in drug theirdesign functions process. are regulated through allosteric Keywords: Allosteric, Activator: Drug design Introduction and ultimately cause disease. While various biological processes expressed the control at different points in life time of protein function is pivotal. As all the cell processes are under carful For the survival of all organisms the significance of protein included regulation of gene expression, translation into protein control and if not properly controls this leads to the abnormality through control of activity and at last degradation of protein [1]. -

Tyrosine Kinase – Role and Significance in Cancer
Int. J. Med. Sci. 2004 1(2): 101-115 101 International Journal of Medical Sciences ISSN 1449-1907 www.medsci.org 2004 1(2):101-115 ©2004 Ivyspring International Publisher. All rights reserved Review Tyrosine kinase – Role and significance in Cancer Received: 2004.3.30 Accepted: 2004.5.15 Manash K. Paul and Anup K. Mukhopadhyay Published:2004.6.01 Department of Biotechnology, National Institute of Pharmaceutical Education and Research, Sector-67, S.A.S Nagar, Mohali, Punjab, India-160062 Abstract Tyrosine kinases are important mediators of the signaling cascade, determining key roles in diverse biological processes like growth, differentiation, metabolism and apoptosis in response to external and internal stimuli. Recent advances have implicated the role of tyrosine kinases in the pathophysiology of cancer. Though their activity is tightly regulated in normal cells, they may acquire transforming functions due to mutation(s), overexpression and autocrine paracrine stimulation, leading to malignancy. Constitutive oncogenic activation in cancer cells can be blocked by selective tyrosine kinase inhibitors and thus considered as a promising approach for innovative genome based therapeutics. The modes of oncogenic activation and the different approaches for tyrosine kinase inhibition, like small molecule inhibitors, monoclonal antibodies, heat shock proteins, immunoconjugates, antisense and peptide drugs are reviewed in light of the important molecules. As angiogenesis is a major event in cancer growth and proliferation, tyrosine kinase inhibitors as a target for anti-angiogenesis can be aptly applied as a new mode of cancer therapy. The review concludes with a discussion on the application of modern techniques and knowledge of the kinome as means to gear up the tyrosine kinase drug discovery process. -

DNA Breakpoint Assay Reveals a Majority of Gross Duplications Occur in Tandem Reducing VUS Classifications in Breast Cancer Predisposition Genes
© American College of Medical Genetics and Genomics ARTICLE Corrected: Correction DNA breakpoint assay reveals a majority of gross duplications occur in tandem reducing VUS classifications in breast cancer predisposition genes Marcy E. Richardson, PhD1, Hansook Chong, PhD1, Wenbo Mu, MS1, Blair R. Conner, MS1, Vickie Hsuan, MS1, Sara Willett, MS1, Stephanie Lam, MS1, Pei Tsai, CGMBS, MB (ASCP)1, Tina Pesaran, MS, CGC1, Adam C. Chamberlin, PhD1, Min-Sun Park, PhD1, Phillip Gray, PhD1, Rachid Karam, MD, PhD1 and Aaron Elliott, PhD1 Purpose: Gross duplications are ambiguous in terms of clinical cohort, while the remainder have unknown tandem status. Among interpretation due to the limitations of the detection methods that the tandem gross duplications that were eligible for reclassification, cannot infer their context, namely, whether they occur in tandem or 95% of them were upgraded to pathogenic. are duplicated and inserted elsewhere in the genome. We Conclusion: DBA is a novel, high-throughput, NGS-based method investigated the proportion of gross duplications occurring in that informs the tandem status, and thereby the classification of, tandem in breast cancer predisposition genes with the intent of gross duplications. This method revealed that most gross duplica- informing their classifications. tions in the investigated genes occurred in tandem and resulted in a Methods: The DNA breakpoint assay (DBA) is a custom, paired- pathogenic classification, which helps to secure the necessary end, next-generation sequencing (NGS) method designed to treatment options for their carriers. capture and detect deep-intronic DNA breakpoints in gross duplications in BRCA1, BRCA2, ATM, CDH1, PALB2, and CHEK2. Genetics in Medicine (2019) 21:683–693; https://doi.org/10.1038/s41436- Results: DBA allowed us to ascertain breakpoints for 44 unique 018-0092-7 gross duplications from 147 probands. -

Integrated Computational Approaches and Tools for Allosteric Drug Discovery
International Journal of Molecular Sciences Review Integrated Computational Approaches and Tools for Allosteric Drug Discovery Olivier Sheik Amamuddy 1 , Wayde Veldman 1 , Colleen Manyumwa 1 , Afrah Khairallah 1 , Steve Agajanian 2, Odeyemi Oluyemi 2, Gennady M. Verkhivker 2,3,* and Özlem Tastan Bishop 1,* 1 Research Unit in Bioinformatics (RUBi), Department of Biochemistry and Microbiology, Rhodes University, Grahamstown 6140, South Africa; [email protected] (O.S.A.); [email protected] (W.V.); [email protected] (C.M.); [email protected] (A.K.) 2 Graduate Program in Computational and Data Sciences, Keck Center for Science and Engineering, Schmid College of Science and Technology, Chapman University, One University Drive, Orange, CA 92866, USA; [email protected] (S.A.); [email protected] (O.O.) 3 Department of Biomedical and Pharmaceutical Sciences, Chapman University School of Pharmacy, Irvine, CA 92618, USA * Correspondence: [email protected] (G.M.V.); [email protected] (Ö.T.B.); Tel.: +714-516-4586 (G.M.V.); +27-46-603-8072 (Ö.T.B.) Received: 25 December 2019; Accepted: 21 January 2020; Published: 28 January 2020 Abstract: Understanding molecular mechanisms underlying the complexity of allosteric regulation in proteins has attracted considerable attention in drug discovery due to the benefits and versatility of allosteric modulators in providing desirable selectivity against protein targets while minimizing toxicity and other side effects. The proliferation of novel computational approaches for predicting ligand–protein interactions and binding using dynamic and network-centric perspectives has led to new insights into allosteric mechanisms and facilitated computer-based discovery of allosteric drugs. Although no absolute method of experimental and in silico allosteric drug/site discovery exists, current methods are still being improved. -

Tyrosine Kinases
KEVANM SHOKAT MINIREVIEW Tyrosine kinases: modular signaling enzymes with tunable specificities Cytoplasmic tyrosine kinases are composed of modular domains; one (SHl) has catalytic activity, the other two (SH2 and SH3) do not. Kinase specificity is largely determined by the binding preferences of the SH2 domain. Attaching the SHl domain to a new SH2 domain, via protein-protein association or mutation, can thus dramatically change kinase function. Chemistry & Biology August 1995, 2:509-514 Protein kinases are one of the largest protein families identified, This is a result of the overlapping substrate identified to date; over 45 new members are identified specificities of many tyrosine kinases, which makes it each year. It is estimated that up to 4 % of vertebrate pro- difficult to dissect the individual signaling pathways by teins are protein kinases [l].The protein kinases are cate- scanning for unique target motifs [2]. gorized by their specificity for serineithreonine, tyrosine, or histidine residues. Protein tyrosine kinases account for The apparent promiscuity of individual tyrosine kinases roughly half of all kinases. They occur as membrane- is a result of their unique structural organization. bound receptors or cytoplasmic proteins and are involved Enzyme specificity is typically programmed by one in a wide variety of cellular functions, including cytokine binding site, which recognizes the substrate and also con- responses, antigen-dependent immune responses, cellular tains exquisitely oriented active-site functional groups transformation by RNA viruses, oncogenesis, regulation that help to lower the energy of the transition state for of the cell cycle, and modification of cell morphology the conversion of specific substrates to products.Tyrosine (Fig. -

Structure and Evolution of Protein Allosteric Sites
Structure and evolution of protein allosteric sites by Alejandro Panjkovich Thesis submitted to Universitat Aut`onoma de Barcelona in partial fulfillment of the requirements for the degree of Doctor of Philosophy Director - Prof. Xavier Daura Tesi Doctoral UAB/ANY 2013 Ph.D. Program - Protein Structure and Function Institut de Biotecnologia i Biomedicina caminante, no hay camino, se hace camino al andar Antonio Machado,1912 Acknowledgements First of all I would like to thank my supervisor and mentor Prof. Xavier Daura for his consistent support and trust in my work throughout these years. Xavi, I deeply appreciate the freedom you gave me to develop this project while you were still carefully aware of the small details. Working under your supervision has been a rich and fulfilling experience. Of course, thanks go as well to current and past members of our institute, especially Rita Rocha, Pau Marc Mu˜noz,Oscar Conchillo, Dr. Mart´ınIndarte, Dr. Mario Ferrer, Prof. Isidre Gibert, Dr. Roman Affentranger and Dr. Juan Cedano for their technical and sometimes philo- sophical assistance. Help from the administrative staff was also significant, I would like to thank in particular Eva, Alicia and Miguel who where always ready to help me in sorting out unexpected bureaucratic affairs. I would also like to thank Dr. Mallur Srivatasan Madhusudhan and his group (especially Kuan Pern Tan, Dr. Minh Nguyen and Binh Nguyen), and also Dr. Gloria Fuentes, Cassio Fernandes, Youssef Zaki, Thijs Kooi, Rama Iyer, Christine Low and many others at the Bioinformatics Institute BII - A∗STAR in Singapore for the many interesting discussions and support during my stage over there. -

Understanding Drug-Drug Interactions Due to Mechanism-Based Inhibition in Clinical Practice
pharmaceutics Review Mechanisms of CYP450 Inhibition: Understanding Drug-Drug Interactions Due to Mechanism-Based Inhibition in Clinical Practice Malavika Deodhar 1, Sweilem B Al Rihani 1 , Meghan J. Arwood 1, Lucy Darakjian 1, Pamela Dow 1 , Jacques Turgeon 1,2 and Veronique Michaud 1,2,* 1 Tabula Rasa HealthCare Precision Pharmacotherapy Research and Development Institute, Orlando, FL 32827, USA; [email protected] (M.D.); [email protected] (S.B.A.R.); [email protected] (M.J.A.); [email protected] (L.D.); [email protected] (P.D.); [email protected] (J.T.) 2 Faculty of Pharmacy, Université de Montréal, Montreal, QC H3C 3J7, Canada * Correspondence: [email protected]; Tel.: +1-856-938-8697 Received: 5 August 2020; Accepted: 31 August 2020; Published: 4 September 2020 Abstract: In an ageing society, polypharmacy has become a major public health and economic issue. Overuse of medications, especially in patients with chronic diseases, carries major health risks. One common consequence of polypharmacy is the increased emergence of adverse drug events, mainly from drug–drug interactions. The majority of currently available drugs are metabolized by CYP450 enzymes. Interactions due to shared CYP450-mediated metabolic pathways for two or more drugs are frequent, especially through reversible or irreversible CYP450 inhibition. The magnitude of these interactions depends on several factors, including varying affinity and concentration of substrates, time delay between the administration of the drugs, and mechanisms of CYP450 inhibition. Various types of CYP450 inhibition (competitive, non-competitive, mechanism-based) have been observed clinically, and interactions of these types require a distinct clinical management strategy. This review focuses on mechanism-based inhibition, which occurs when a substrate forms a reactive intermediate, creating a stable enzyme–intermediate complex that irreversibly reduces enzyme activity. -

Spring 2013 Lecture 13-14
CHM333 LECTURE 13 – 14: 2/13 – 15/13 SPRING 2013 Professor Christine Hrycyna INTRODUCTION TO ENZYMES • Enzymes are usually proteins (some RNA) • In general, names end with suffix “ase” • Enzymes are catalysts – increase the rate of a reaction – not consumed by the reaction – act repeatedly to increase the rate of reactions – Enzymes are often very “specific” – promote only 1 particular reaction – Reactants also called “substrates” of enzyme catalyst rate enhancement non-enzymatic (Pd) 102-104 fold enzymatic up to 1020 fold • How much is 1020 fold? catalyst time for reaction yes 1 second no 3 x 1012 years • 3 x 1012 years is 500 times the age of the earth! Carbonic Anhydrase Tissues ! + CO2 +H2O HCO3− +H "Lungs and Kidney 107 rate enhancement Facilitates the transfer of carbon dioxide from tissues to blood and from blood to alveolar air Many enzyme names end in –ase 89 CHM333 LECTURE 13 – 14: 2/13 – 15/13 SPRING 2013 Professor Christine Hrycyna Why Enzymes? • Accelerate and control the rates of vitally important biochemical reactions • Greater reaction specificity • Milder reaction conditions • Capacity for regulation • Enzymes are the agents of metabolic function. • Metabolites have many potential pathways • Enzymes make the desired one most favorable • Enzymes are necessary for life to exist – otherwise reactions would occur too slowly for a metabolizing organis • Enzymes DO NOT change the equilibrium constant of a reaction (accelerates the rates of the forward and reverse reactions equally) • Enzymes DO NOT alter the standard free energy change, (ΔG°) of a reaction 1. ΔG° = amount of energy consumed or liberated in the reaction 2. -

Structural Basis for Regiospecific Midazolam Oxidation by Human Cytochrome P450 3A4
Structural basis for regiospecific midazolam oxidation by human cytochrome P450 3A4 Irina F. Sevrioukovaa,1 and Thomas L. Poulosa,b,c aDepartment of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900; bDepartment of Chemistry, University of California, Irvine, CA 92697-3900; and cDepartment of Pharmaceutical Sciences, University of California, Irvine, CA 92697-3900 Edited by Michael A. Marletta, University of California, Berkeley, CA, and approved November 30, 2016 (received for review September 28, 2016) Human cytochrome P450 3A4 (CYP3A4) is a major hepatic and Here we describe the cocrystal structure of CYP3A4 with the intestinal enzyme that oxidizes more than 60% of administered drug midazolam (MDZ) bound in a productive mode. MDZ therapeutics. Knowledge of how CYP3A4 adjusts and reshapes the (Fig. 1) is the benzodiazepine most frequently used for pro- active site to regioselectively oxidize chemically diverse com- cedural sedation (9) and is a marker substrate for the CYP3A pounds is critical for better understanding structure–function rela- family of enzymes that includes CYP3A4 (10). MDZ is hydrox- tions in this important enzyme, improving the outcomes for drug ylated by CYP3A4 predominantly at the C1 position, whereas metabolism predictions, and developing pharmaceuticals that the 4-OH product accumulates at high substrate concentrations have a decreased ability to undergo metabolism and cause detri- (up to 50% of total product) and inhibits the 1-OH metabolic mental drug–drug interactions. However, there is very limited pathway (11–14). The reaction rate and product distribution structural information on CYP3A4–substrate interactions available strongly depend on the assay conditions and can be modulated by to date. -
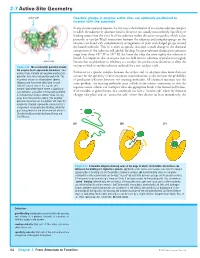
2-7 Active-Site Geometry
2-7 Active-Site Geometry active site Reactive groups in enzyme active sites are optimally positioned to interact with the substrate In any enzyme-catalyzed reaction, the first step is the formation of an enzyme–substrate complex in which the substrate or substrates bind to the active site, usually noncovalently. Specificity of binding comes from the close fit of the substrate within the active-site pocket, which is due primarily to van der Waals interactions between the substrate and nonpolar groups on the enzyme, combined with complementary arrangements of polar and charged groups around the bound molecule. This fit is often so specific that even a small change in the chemical composition of the substrate will abolish binding. Enzyme–substrate dissociation constants range from about 10–3 M to 10–9 M; the lower the value the more tightly the substrate is bound. It is important that an enzyme does not hold onto its substrates or products too tightly active site because that would reduce its efficiency as a catalyst: the product must dissociate to allow the Figure 2-22 The electrostatic potential around enzyme to bind to another substrate molecule for a new catalytic cycle. the enzyme Cu,Zn-superoxide dismutase Red contour lines indicate net negative electrostatic Formation of a specific complex between the catalyst and its substrates does more than just potential; blue lines net positive potential. The account for the specificity of most enzymatic transformations: it also increases the probability enzyme is shown as a homodimer (green of productive collisions between two reacting molecules. All chemical reactions face the ribbons) and two active sites (one in each same problem: the reacting molecules must collide in the correct orientation so that the subunit) can be seen at the top left and bottom right of the figure where a significant requisite atomic orbitals can overlap to allow the appropriate bonds to be formed and broken. -

Download 20190410); Fragmentation for 20 S
ARTICLE https://doi.org/10.1038/s41467-020-17387-y OPEN Multi-layered proteomic analyses decode compositional and functional effects of cancer mutations on kinase complexes ✉ Martin Mehnert 1 , Rodolfo Ciuffa1, Fabian Frommelt 1, Federico Uliana1, Audrey van Drogen1, ✉ ✉ Kilian Ruminski1,3, Matthias Gstaiger1 & Ruedi Aebersold 1,2 fi 1234567890():,; Rapidly increasing availability of genomic data and ensuing identi cation of disease asso- ciated mutations allows for an unbiased insight into genetic drivers of disease development. However, determination of molecular mechanisms by which individual genomic changes affect biochemical processes remains a major challenge. Here, we develop a multilayered proteomic workflow to explore how genetic lesions modulate the proteome and are trans- lated into molecular phenotypes. Using this workflow we determine how expression of a panel of disease-associated mutations in the Dyrk2 protein kinase alter the composition, topology and activity of this kinase complex as well as the phosphoproteomic state of the cell. The data show that altered protein-protein interactions caused by the mutations are asso- ciated with topological changes and affected phosphorylation of known cancer driver pro- teins, thus linking Dyrk2 mutations with cancer-related biochemical processes. Overall, we discover multiple mutation-specific functionally relevant changes, thus highlighting the extensive plasticity of molecular responses to genetic lesions. 1 Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, -

Regulation of the Stability of the Protein Kinase DYRK1A: Establishing Connections with the Wnt Signaling Pathway
Regulation of the stability of the protein kinase DYRK1A: establishing connections with the Wnt signaling pathway Krisztina Arató TESI DOCTORAL UPF / 2010 Barcelona, November 2010 Regulation of the stability of the protein kinase DYRK1A: establishing connections with the Wnt signaling pathway Krisztina Arató Memòria presentada per optar al grau de Doctora per la Universitat Pompeu Fabra. Aquesta tesi ha estat realitzada sota la direcció de la Dra. Susana de la Luna al Centre de Regulació Genòmica (CRG, Barcelona), dins del Programa de Genes i Malaltia. Krisztina Arató Susana de la Luna A Pere, por haberme traído a Barcelona… Cover design by Luisa Lente (www.yoyo.es). Index Page Abstract/Resumen.................................................................................. 1 Introduction............................................................................................. 5 The protein kinase DYRK1A................................................................ 7 The DYRK family of protein ....................................................... 7 Structure and mechanism of activation of DYRK1A kinase........ 8 Regulation of DYRK1A expression ............................................ 10 Regulation of DYRK1A subcellular localization.......................... 11 Regulation of DYRK1A activity................................................... 13 DYRK1A as a regulator of signaling pathways........................... 14 The Notch signaling pathway........................................... 16 Receptor tyrosine kinase signaling.................................