(Hemochromogen Or
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterisation, Classification and Conformational Variability Of
Characterisation, Classification and Conformational Variability of Organic Enzyme Cofactors Julia D. Fischer European Bioinformatics Institute Clare Hall College University of Cambridge A thesis submitted for the degree of Doctor of Philosophy 11 April 2011 This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. This dissertation does not exceed the word limit of 60,000 words. Acknowledgements I would like to thank all the members of the Thornton research group for their constant interest in my work, their continuous willingness to answer my academic questions, and for their company during my time at the EBI. This includes Saumya Kumar, Sergio Martinez Cuesta, Matthias Ziehm, Dr. Daniela Wieser, Dr. Xun Li, Dr. Irene Pa- patheodorou, Dr. Pedro Ballester, Dr. Abdullah Kahraman, Dr. Rafael Najmanovich, Dr. Tjaart de Beer, Dr. Syed Asad Rahman, Dr. Nicholas Furnham, Dr. Roman Laskowski and Dr. Gemma Holli- day. Special thanks to Asad for allowing me to use early development versions of his SMSD software and for help and advice with the KEGG API installation, to Roman for knowing where to find all kinds of data, to Dani for help with R scripts, to Nick for letting me use his E.C. tree program, to Tjaart for python advice and especially to Gemma for her constant advice and feedback on my work in all aspects, in particular the chemistry side. Most importantly, I would like to thank Prof. Janet Thornton for giving me the chance to work on this project, for all the time she spent in meetings with me and reading my work, for sharing her seemingly limitless knowledge and enthusiasm about the fascinating world of enzymes, and for being such an experienced and motivational advisor. -

Simple Proteins Conjugated Proteins Enzymes
Ministry of Health of Ukraine Zaporizhzhia State Medical University Biochemistry & Laboratory Diagnostics Department Simple proteins Conjugated proteins Enzymes A manual for study of submodule 1 on Biochemistry for second-year students of International Faculty specialty: 7.12010001 ―General medicine‖ Zaporizhzhia-2015 This manual is recommended as additional one for independent work of students at home and in class for submodule 1 of Biochemistry at 2-d year study at specialty 7.12010001 ―General Medicine‖. Editors: Dr.Hab. professor Aleksandrova K.V. PhD, associate professor Krisanova N.V. PhD, assistant Levich S.V. Rewiewers Head of Department of Organic and Bioorganic Chemistry ZSMU, Dr.Hab. professor Kovalenko S. I. Head of Department of Biology, Parasitology and Genetics ZSMU, Dr.Hab. associate professor Pryhod’ko O. B. 2 Вступ Наведений навчально-методичний посібник пропонується викладачами кафедри біохімії та лабораторної діагностики для використання у самостійній роботі англомовних іноземних студентів медичного факультету ЗДМУ з метою підготовки до змістового модулю 1 з біохімії. Цей навчальний посібник містить все необхідне для вивчення базових питань зi змістового модулю 1 «Прості та складні білки. Ферменти» відповідно робочої програми з дисципліни «Біохімія» для студентів 2-го курсу медичного факультету вищих навчальних медичних закладів III-IV рівнів акредитації в Україні. Автори INTRODUCTION This methodological manual is recommended for students to use like additional materials in preparation for practical classes on Biochemistry for submodule 1. The plan to work with it: 1) to prepare theoretical questions for topic using literature sources, summaries of lectures and this textbook; 2) to make testing control yourself; 3) to be ready to answer teacher about principles of methods used for the determination of proposed biochemical indexes and about their clinical significance. -

BLOOD-2,3 Done By
BLOOD-2,3 أ آ ☺ ه اآة ه رة آ ا ا ات اآر + MCQ + ات ﻡ اب وأام ﺏ Its like blood juice ☺ Done by أ ا Special thanks for abo yosra I appreciate ur great help & Thanx for Hegaz group I add some notes from them 1 BLOOD-2,3 BLOOD CELLS : • ◙ Erythrocytes (Red blood cells) • Leukocytes (White blood cells) – Granulocytes • Neutrophils • Basophils • Eosinophils – Monocytes – Lymphocytes • T • B • Megacaryocyte (Platelets) RBC • Men 4.6 – 6.2 x 10¹²/l. • Women 4.2 – 5.4 x 10¹²/l • Total number of red cells in circulation = 2.5x1013 WBC • Men and women 5-7 x 109/l. Platelets • Men and women 250 x 109/l Hb • Men 14 – 16 g/l • Women 12 – 16 g/l PCV (Haematocrit) • Men 0.42 – 0.52 l/l • Women 0.37 – 0.47 l/l * RBC : ••• Biconcave disks: Highly specialized - Diameter 6 - 9 µm - Thickness: 1 - 2 µm - Volume; ~ 88 fl. ••• Deformable - i.e. can change shape to transverse smallest blood vessels. ••• Contain haemoglobin (~ 33%). (MCQ) ••• No nucleus or mitochondria. ••• Function: Transport of O2 and CO2. ••• Normal Range: - 5.5 ±±± 1.0 x 1012/L - 4.8 ±±± 1.0 x 1012/L 2 BLOOD-2,3 • Deliver oxygen to tissues and CO2 from tissues to lungs. • Red cell life span is 120 days. • Senescent red cells are destroyed by spleen and replaced by juvenile cells released by bone marrow. • An average 70 Kg adult male produces 2.3x106 red cell/sec. ERYTHROCYTE STRUCTURE • Biconcave shape. Spherical. • Simple structure: – Membrane surrounding cytoplasm. – Almost 95% of solutes in cytosal is haemoglobin. -

The Classification of Proteins
THE CLASSIFICATION OF PROTEINS - According to their function - 1. As catalysts, i.e. enzymes. 2. As structural elements (Collagen, Elastin). 3. As mode of transport (Albumin, Globulin, Hemoglobin) 4. As hormones (Insulin, Growth hormones). 5. As protective agents (Antibodies, Blood clothing) 6. As contractive elements (Actin, Myosin) - According to their composition - 1. Simple proteins. Simple proteins are made up of amino acids only and on hydrolysis yield amino acid mixture only. 2. Conjugated Proteins They are proteins that contain a non-protein group (also ‘prosthetic group’) attached to the protein part. It has a non-protein component and amino acid mixture. Conjugated Protein = Protein part + Prosthetic group . Conjugated proteins are classified according to the nature of the non-protein group attached to the protein part ( Ошибка! Источник ссылки не найден. ). Exsample of simple proteins 1. Fibrous proteins a. Collagen It contains high proportion of hydroxyproline and hydroxylysine. It is a major protein of connective tissues. On boiling with water it forms gelatin. b. Elastin It is present in tendons and arteries. с. Keratin It contains large amount of sulphur as cystine. It is present in hair, wool, nails etc. 2. Globular Proteins a. Albumins Serum albumin and ovalbumin of egg white. It is water-soluble. It is precipitated from solution by full saturation of ammonium sulphate. It is coagulated by heat. b. Globulins Serum globulins, fibrinogens and muscle myosin. It is soluble in dilute salt solutions. It is precipitated from solution by half saturation of ammonium sulphate. It is coagulated by heat. c. Glutelins Cereal proteins such as glutelins of wheat, oxyzenin from rice and zein of maize. -

Peptide Bond
Peptide bond Peptide bond: A peptide bond is an amide type of covalent chemical bond formed between two amino acids when the carboxyl group of one amino acid reacts with the amino group of the other amino acid. Peptide : A linear chain of amino acid residues held together by peptide bond. Dipeptide: Consists of 2 amino acids. Tripeptide: Consists of 3 amino acids. Oligopeptide: Consists of 4-10 amino acids. Polypeptide: Consists of more than 50 amino acids. Poll Question-01 Oligopeptide consists of ___ (a) 3-10 amino acids (b) 4-10 amino acids (c) 50 amino acids (d) More than 50 amino acids Proteins Proteins are large organic compounds composed of numerous amino acids. A protein molecule is composed of 100 or more amino acid molecules. Classification of Protein Protein Based on Based on Based on Based on chemical Based on biological shape structure properties & solubility nutritional value activities 1. Structural 1. Fibrous 1. Primary 1. Simple 1. 1st class 2. Functional 2. Globular 2. Secondary 2. Conjugated 2. 2nd class 3. Tertiary 3. Derived 4. Quaternary Classification of protein based on biological activities Structural protein Keratin, collagen, fibrin, sclerotin, chondrin, sein Functional protein Enzyme, hormone, vitamins, respiratory pigments Classification of protein based on shape Fibrous protein: Globular protein: Keratin, collagen, Myoglobin, Insulin, fibrin. Hemoglobin. Classification of protein based on chemical properties & solubility Simple protein Conjugated protein Derived protein 1. Albumin 1. Nucleoprotein 1. Myosan 2. Globulin 2. Glycoprotein 2. Albumose 3. Glutelin 3. Lipoprotein 4. Prolamin 4. Chromoprotein 5. Histone 5. Metalloprotien 6. Protamine 6. Phosphoprotein 7. -
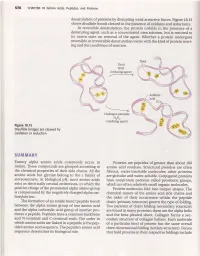
Swffiffe&Ffiy
576 CHAPTERl8 Amino Acids,Peptides, and Proteins denaturation of proteins by disrupting weak attractive forces. Figure lB.t5 shows disulfide bonds cleaved in the presenceof oxidants and reductants. In reversible denaturation, the protein unfolds in the presence of a denaturing agent, such as a concentrated urea solution, but is restored to its native state on removal of the agent. \Mhether a protein undergoes reversible or irreversible denaturation varies with the kind of protein react- ing and the conditions of reaction. Thiol RSH (reducing agent) Hydrogen peroxide Hroz (oxidizing agent) Figure18.15 Disulfidebridges are cleaved by oxidationor reduction SWffifFE&ffiY Twenty alpha amino acids commonly occur in Proteins are peptides of greaterthan about 100 nature. Thesecompounds are grouped accordingto amino acid residues.Structural proteins are often the chemical properties of their side chains. All the fibrous, water-insoluble molecules; other proteins amino acids but glycine belong to the I family of are globular and water-soluble.Conjugated proteins stereoisomers.At biological pH, most amino acids have nonprotein portions called prosthetic groups, exist as electrically neutral zwitterions, in which the which are often relatively small organic molecules. positive charge of the protonated alpha amino group Protein molecules fold into unique shapes.The is compensated by the negatively charged alpha car- chemical nature of the amino acid side chains and boxylate ion. the order of their occurrence within the peptide The formation of an amide bond (peptidebond) chain (primary structure) govern the type of folding. between the alpha amino group of one amino acid Two patterns of chain folding (secondary structure) and the alpha carboxylic acid group of another pro- are found in many proteins; these are the alpha helix ducesa peptide. -

Structural and Functional Studies of Hemoproteins
STRUCTURAL AND FUNCTIONAL STUDIES OF HEMOPROTEINS FROM POLAR MARINE ORGANISMS Daniela Coppola Dottorato in Scienze Biotecnologiche – XXIV ciclo Indirizzo Biotecnologie Industriale e Molecolare Università di Napoli Federico II Dottorato in Scienze Biotecnologiche – XXIV ciclo Indirizzo Biotecnologie Industriale e Molecolare Università di Napoli Federico II STRUCTURAL AND FUNCTIONAL STUDIES OF HEMOPROTEINS FROM POLAR MARINE ORGANISMS Daniela Coppola Dottoranda: Daniela Coppola Relatore: Prof. Lelio Mazzarella Prof. Giovanni Sannia Co-tutore Dr. Cinzia Verde Coordinatore: Prof. Giovanni Sannia Ai miei genitori, mia sorella Sabina e Lorenzo perché non sarei la persona che sono oggi senza il loro amore ed il loro costante sostegno. INDEX ABBREVIATIONS 3 RIASSUNTO 5 ABSTRACT 11 CHAPTER I: INTRODUCTION 1.1 Antarctic marine organisms and their evolutionary adaptations 13 1.2 Vertebrate hemoglobins 16 1.2.1 Hemoglobins in suborder Notothenioidei 18 1.2.2 Hemoglobin-based blood substitutes 21 1.3 Bacterial globins 23 1.3.1 Globins of Pseudoalteromonas haloplanktis TAC125 and their involvement in oxidative and nitrosative stress 25 1.3.2 Impact of the bacterial hemoglobins in biotechnology 27 1.4 Objectives of the PhD project 28 CHAPTER II: STRUCTURAL AND FUNCTIONAL CHARACTERISATION OF HIGH- AND SUB-ANTARCTIC NOTOTHENIOID HEMOGLOBINS 2.1 Introduction 29 2.2 Results and discussion 29 2.2.1 Purification of Hbs and separation of globins 29 2.2.2 Separation of globins and primary structure 30 2.2.3 Oxygen-binding properties 32 2.2.4 Kinetics of oxygen -

Ii Christopher Elliot Tubbs Committee Chair, Joseph C. Hall, Ph.D. Using
ii ABSTRACT THE BIOCHEMICAL CHARACTERIZATION OF PROTEIN DE AND ITS INTERACTION WITH RAT EPIDIDYMAL SPERM. Christopher Elliot Tubbs Committee Chair, Joseph C. Hall, Ph.D. Using traditional column chromatography, Protein DE has been purified from rat epididymides. Affinity, size exclusion, and ion-exchange chromatography were utilized to purify the protein to homogeneity. Protein DE purity was demonstrated using one and two-dimensional electrophoresis. Using the purified sample, an accurate molecular mass of 27,534 Daltons was determined using electrospray-ionization mass spectrometry. After four chromatographic steps, Protein DE was efficiently separated from all detectable epididymal proteins. This report provides the first rapid and reproducible method for purifying protein DE to homogeneity. Using western blot analysis and immunofluorescence, protein D is initially detected in rat epididymal tissue and associated with sperm from the distal caput region. In contrast, when sperm were recovered from the female reproductive tract seven hours after mating, protein D was not detected by western blot, but did display faint immunofluorescence. Additionally, using photoactivatible cross-linking, a 120 KD sperm membrane protein that specifically interacts with protein D was identified. A population of membrane bound protein D was released from NaCl washed epididymal sperm when incubated in the presence of phosphatidyl-inositol specific phospholipase C. This report is the first demonstrating that both the secretion and sperm-association of protein D occur in the distal caput region of the rat epididymis. It is the only report showing western blot iii analysis and immunolocalization of sperm-associated protein D on sperm deposited in the female reproductive tract after mating. -

(April 2018) Model Answers Chemistry I- Biorganic Chemistry
QP Code: 3834 F.Y.B.Sc. in Biotechnology Semester II Examination (April 2018) Model Answers Chemistry I- Biorganic chemistry Q.1. Do as directed: (Any fifteen) 15 Define the following terms: 1) Carbohydrates Carbohydrates are poly hydroxy aldehydes or ketones with a general formula Cn(H2O)n, where n 3 2) Dipeptide A dipeptide holds two interacting amino acids with one peptide bond 3) Isoelectric pH At isoelectric pH the amino acids exists as a zwitter ions and they bear no net charge. 4) Isomers Isomers are the compounds having the same empirical formula and the structure, but they differ from one another by a configuration around only one carbon atom. 5) Unsaturated fatty acid Unsaturated Fatty acids are carboxylic acids with atleast one or more double bonds in its hydrocarbon chain. 6) Pitch of the helix- is the height of one complete helix turn, measured parallel to the axis of the helix. 7) Saponification When animal fat/oil reacts with aqueous NaOH or KOH, they form soap and glycerol. Since this reaction leads to the formation of soap, it is called the Saponification. Draw structures of the following biomolecules: 8) Purine base. Any one base 9) D- glyceraldehydes CHO-CHOH-CH2OH 10) D-glucose 11) Serine 12) Acetone Name the following: 13) An acidic amino acid- Aspartic acid, glutamic acid 14) One keto sugar- Fructose, ribulose 15) Sterol of biological significance - cholesterol 16) Fibrous protein present in hair - Keratin 17) Sugar present in DNA. -deoxyribose 18) Parent compound of phospholipids- Phosphatidic acid 19) The transport protein present in the red blood cell - Albumin 20) Type of bond present between base pair G-C – Hydrogen bond Q2. -

Amino Acids and Protein DEFINITION of AMINO ACID Organic Compounds Contains N in Addition to C , H, O
Amino Acids And Protein DEFINITION OF AMINO ACID Organic compounds Contains N in addition to C , H, O Contains at least two functional groups -COOH and -NH2 attached to ALPHA –C atom They are building blocks of proteins Essential for life GENERAL STRUCTURE OF Α - AMINO ACID Reference axis COOH H H2N C R Side chain defines 20 amino acids L- α Amino acid STRUCTURE OF Α - AMINO ACID WHAT IS D & L FORM COOH CHO COOH H H H NH2 H2N C C OH C R R CH2OH L- Amino acid D- Glyceraldehyde D- Amino acid TYPES OF AMINO ACIDS α Amino acids --- Natural amino acids β – Amino acids -- β alanine, β taurine γ -- Amino acids --- GABA, L Amino acids - normal present in Human D Amino acids - present in Bacteria and virus FUNCTIONS OF AMINO ACIDS 1. Components of peptides and proteins 2. Derivatives are components of lipids 3. Neurotransmitters or precursors of NT or hormones 4. Glucogenic 5. Purine and pyrimidine Nitrogen base synthesis FUNCTIONS OF AMINO ACIDS 6. Non-proteinogenic AA are intermediate of metabolic reactions 7. Forms active site in Enzymes 8. Acts as buffers at physiological pH -- Histidine 9. Transport oxygen and Carbon dioxide -- Histidine 10. Detoxification agents -- Glycine, Cysteine, Taurine VARIOUS WAYS OF CLASSIFICATION 1. Based on their presence in protein 2. Based on R Group (side chain) 3. Based on Nutritional value 4. Based on metabolic fate 5. Based on polarity of side chain CLASSIFICATION BASED ON R GROUP (SIDE CHAIN) COOH COOH COOH H2N C H H2N C H H2N C H H CH3 R BASED ON R GROUP / SIDE CHAIN A A = Aliphatic amino acids S S = Sulphur -

Chemistry and Functions of Hemoglobin and Myoglobin
Chemistry And Functions Of Hemoglobin and Myoglobin Dr Bela Goyal • What are Hemoproteins? • What is Hemoglobin? • Structure of Hemoglobin • Functions of Hemoglobin • ODC and Factors affecting it • Normal Hb Variants • Hemoglobin Derivatives • Hemoglobinopathies Learning objectives What are Hemoproteins? What is Hemoglobin? Structure of Hemoglobin What Are Hemoproteins? • Hemoproteins are Conjugated Proteins • With Heme as a Prosthetic group in their structures. Heme Containing Proteins 1. Hemoglobin (Hb) 2. Myoglobin (Mb) 3. Cytochromes (ETC Components) Heme Containing Enzymes 1. Catalase 2. Peroxidase 3. Tryptophan Dioxygenase/ Tryptophan Pyrrolase What Is Hemoglobin? • Hemoglobin(Hb) is a major Hemoprotein of Human body • Hemoglobin Chemically is: • Conjugated Protein • In Hemoglobin • Heme is a Prosthetic group • Globin is a Protein part (Hemoglobin = Heme + Globin) • Hemoglobin(Hb) is Red color pigment • Location Of Hemoglobin- Inside Red blood cells/Erythrocytes of blood. • Hemoglobin In RBCs Occupies: • 33% of the RBC volume (1/3) • 90-95% of the dry weight of RBC is by Hb. • Normal concentration of Hemoglobin in the Human Blood: Adult Males- 13.5–17.5 gm/dL Adult Females- 12.5–16.5 gm/dL Terminologies of Hemoglobin • Hemoprotein -Heme is a prosthetic group • Chromoprotein - Red in color • Metalloprotein - Metal Iron (Fe) present • Respiratory Protein- Connected to Respiration process and Respiratory Chain(Electron Transport Chain) • Oxygen Binding Protein-Binds with molecular Oxygen and transports it. Structure of hemoglobin • Iron containing pigment heme is attached to protein globin • Heme is iron porphyrin complex called Iron protoporphyrin IX • Globin is a protein Iron in Heme • Functional form Iron in Heme is- • Ferrous form(Fe++) • Reduced state • Fe++ located centrally in Protoporphyrin ring system. -

Bonds Stabilizing Protein Structure, Levels of Organization in Proteins, Denaturation, Introduction to Simple and Conjugate Proteins
CBCS 3RD SEM (MAJOR) Credits: 4 Unit 3 (II): Proteins BONDS STABILIZING PROTEIN STRUCTURE, LEVELS OF ORGANIZATION IN PROTEINS, DENATURATION, INTRODUCTION TO SIMPLE AND CONJUGATE PROTEINS By- Dr. Luna Phukan Definition Proteins are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells, and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. Protein Bonds Proteins are the polymers of amino acids. Amino acids are joined together by a special type of covalent bond (peptide bond) to form linear structures called polypeptides. The polypeptides are then folded into specific structures to form the functional conformation of the protein. The folding of proteins into specific shapes and conformations are assisted and stabilized by many types of bonds in them. Some of these bonds are strong bonds whereas others are weak interactions. Important types of bonds involved in protein structure and conformation are Peptide bonds, Ionic bonds, Disulfide bonds, Hydrogen bonds and Hydrophobic Interactions. Bonds Stabilizing Protein structure Types of bonds There are 5 types of chemical bonds that play important roles in determining and stabilizing 3-D protein structure. They are: 1) Peptide bonds 2) Ionic Bonds 3) Disulfide Bonds 4) Hydrogen bonds 5) Hydrophobic bonds Peptide Bonds Peptide bond definition: a covalent bond formed between the carboxylic group of one amino acid and the group of another amino acid Peptide bond is a strong covalent bond with high bond dissociation energy.