Investigation of the Embryonic Chicken Pineal Gland As a Model for Experimental Jet Lag
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guthrie Cdna Resource Center
cDNA Resource Center cDNA Resource Center Catalog cDNA Resource Center Missouri University of Science and Technology 400 W 11th Rolla, MO 65409 TEL: (573) 341-7610 FAX: (573) 341-7609 EMAIL: [email protected] www.cdna.org September, 2008 1 cDNA Resource Center Visit our web site for product updates 2 cDNA Resource Center The cDNA Resource Center The cDNA Resource Center is a service provided by the faculty of the Department of Biological Sciences of Missouri University of Science and Technology. The purpose of the cDNA Resource Center is to further scientific investigation by providing cDNA clones of human proteins involved in signal transduction processes. This is achieved by providing high quality clones for important signaling proteins in a timely manner. By high quality, we mean that the clones are • Sequence verified • Propagated in a versatile vector useful in bacterial and mammalian systems • Free of extraneous 3' and 5' untranslated regions • Expression verified (in most cases) by coupled in vitro transcription/translation assays • Available in wild-type, epitope-tagged and common mutant forms (e.g., constitutively- active or dominant negative) By timely, we mean that the clones are • Usually shipped within a day from when you place your order. Clones can be ordered from our web pages, by FAX or by phone. Within the United States, clones are shipped by overnight courier (FedEx); international orders are shipped International Priority (FedEx). The clones are supplied for research purposes only. Details on use of the material are included on the Material Transfer Agreement (page 3). Clones are distributed by agreement in Invitrogen's pcDNA3.1+ vector. -

An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors
Ecology and Evolutionary Biology 2021; 6(3): 53-77 http://www.sciencepublishinggroup.com/j/eeb doi: 10.11648/j.eeb.20210603.11 ISSN: 2575-3789 (Print); ISSN: 2575-3762 (Online) An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors Miguel Angel Fuertes*, Carlos Alonso Department of Microbiology, Centre for Molecular Biology “Severo Ochoa”, Spanish National Research Council and Autonomous University, Madrid, Spain Email address: *Corresponding author To cite this article: Miguel Angel Fuertes, Carlos Alonso. An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors. Ecology and Evolutionary Biology. Vol. 6, No. 3, 2021, pp. 53-77. doi: 10.11648/j.eeb.20210603.11 Received: April 24, 2021; Accepted: May 11, 2021; Published: July 13, 2021 Abstract: Capturing conserved patterns in genes and proteins is important for inferring phenotype prediction and evolutionary analysis. The study is focused on the conserved patterns of the G protein-coupled receptors, an important superfamily of receptors. Olfactory receptors represent more than 2% of our genome and constitute the largest family of G protein-coupled receptors, a key class of drug targets. As no crystallographic structures are available, mechanistic studies rely on the use of molecular dynamic modelling combined with site-directed mutagenesis data. In this paper, we hypothesized that human-mouse orthologs coding for G protein-coupled receptors maintain, at speciation events, shared compositional structures independent, to some extent, of their percent identity as reveals a method based in the categorization of nucleotide triplets by their gross composition. The data support the consistency of the hypothesis, showing in ortholog G protein-coupled receptors the presence of emergent shared compositional structures preserved at speciation events. -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Detection of Polymorphisms in the MTNR1A Gene and Their Association with Reproductive Performance in Awassi Ewes
animals Article Detection of Polymorphisms in the MTNR1A Gene and Their Association with Reproductive Performance in Awassi Ewes Giovanni Cosso 1, Michella Nehme 2, Sebastiano Luridiana 1 , Luisa Pulinas 1, Giulio Curone 3 , Chadi Hosri 4, Vincenzo Carcangiu 1 and Maria Consuelo Mura 1,* 1 Department of Veterinary Medicine of Sassari, University of Sassari, Via Vienna 2, 07100 Sassari, Italy; [email protected] (G.C.); [email protected] (S.L.); [email protected] (L.P.); [email protected] (V.C.) 2 Department of Agriculture and Food Engineering, Faculty of Engineering, Holy Spirit University of Kaslik, Kaslik, Jounieh 446, Lebanon; [email protected] 3 Department of Veterinary Medicine of Milan, University of Milan, Via dell’Università 6, 26900 Lodi, Italy; [email protected] 4 Department of Veterinary Sciences, Faculty of Agriculture, Lebanese University, Dekwaneh, Beirut 14/6573, Lebanon; [email protected] * Correspondence: [email protected]; Tel.: +39-079-229-437; Fax: +39-079-229-592 Simple Summary: The purpose of the study was to explore the influence of MTNR1A gene polymor- phisms on the reproductive performance in Awassi sheep, which is an important and widespread breed in developing Mediterranean countries. A total of 31 SNPs was detected, 5 of which caused amino acid changes. Two of the found SNPs were found to be totally linked and associated with an advanced reproductive recovery in ewes carrying the C allele. The obtained results could be useful for improving reproductive management in developing Mediterranean areas. Citation: Cosso, G.; Nehme, M.; Abstract: The economy in Mediterranean areas is tightly linked to the evolution of the sheep-farming Luridiana, S.; Pulinas, L.; Curone, G.; system; therefore, improvement in ewe’s reproductive performance is essential in the developing Hosri, C.; Carcangiu, V.; Mura, M.C. -

The Evolutionarily Conserved Role of Melatonin in CNS Disorders and Behavioral Regulation Translational Lessons from Zebrafish
Neuroscience and Biobehavioral Reviews 99 (2019) 117–127 Contents lists available at ScienceDirect Neuroscience and Biobehavioral Reviews journal homepage: www.elsevier.com/locate/neubiorev Review article The evolutionarily conserved role of melatonin in CNS disorders and T behavioral regulation: Translational lessons from zebrafish Rafael Genarioa,1, Ana C.V.V. Giacominia,b,1, Konstantin A. Demind,f, Bruna E. dos Santosa, Natalia I. Marchioria, Angrey D. Volging, Alim Bashirzadeg,h, Tamara G. Amstislavskayag,h, ⁎ ⁎⁎ Murilo S. de Abreua,c, , Allan V. Kalueffe,f,g,i,j,k, a Bioscience Institute, University of Passo Fundo (UPF), Passo Fundo, RS, Brazil b Postgraduate Program in Environmental Sciences, University of Passo Fundo (UPF), Passo Fundo, Brazil c The International Zebrafish Neuroscience Research Consortium (ZNRC), Slidell, LA,USA d Institute of Experimental Medicine, Almazov National Medical Research Centre, Ministry of Healthcare of Russian Federation, St. Petersburg, Russia e School of Pharmacy, Southwest University, Chongqing, China f Institute of Translational Biomedicine, St. Petersburg State University, St. Petersburg, Russia g Laboratory of Translational Biopsychiatry, Research Institute of Physiology and Basic Medicine, Novosibirsk, Russia h Department of Neuroscience, Novosibirsk State University, Novosibirsk, Russia i Ural Federal University, Ekaterinburg, Russia j Granov’s Russian Scientific Research Center of Radiology and Surgical Technologies, Ministry of Healthcare of Russian Federation, Pesochny, Russia k Almazov National Medical Research Centre, Ministry of Healthcare of Russian Federation, St. Petersburg, Russia ARTICLE INFO ABSTRACT Keywords: Melatonin is an important hormone regulating circadian rhythm, neuroprotection and neuroimmune processes. Melatonin However, its exact physiological roles in brain mechanisms remain poorly understood. Here, we summarize the Animal model mounting evidence implicating melatonin in brain disorders and behavior, based on clinical and experimental Genetic model studies in-vivo. -
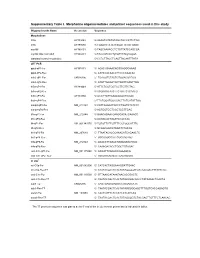
Supplementary Table I. Morpholino Oligonucleotides and Primer Sequences Used in This Study
Supplementary Table I. Morpholino oligonucleotides and primer sequences used in this study Oligonucleotide Name Accession Sequence Morpholinos tlr5a AY389449 5'-AAAGTGTATGTAGCTGCCATTCTGG tlr5b AY389450 5'-TGAATGTATATCCCATTCTGTGAGC myd88 AY388401 5'-TAGCAAAACCTCTGTTATCCAGCGA myd88 5bp mismatch AY388401 5'-TAcCAtAACCTgTGTTATCgAGgGA standard control morpholino 5'-CCTCTTACCTCAGTTACAATTTATA qRT-PCR ppial-qP1-Fw AY391451 5’- ACACTGAAACACGGAGGCAAAG ppial-qP2-Rev 5’- CATCCACAACCTTCCCGAACAC irak3-qP1-Fw CK026195 5’- TGAGGTCTACTGTGGACGATGG irak3-qP2-Rev 5’- ATGTTAGGATGCTGGTTGAGTTGG tlr5a-qP1-Fw AY389449 5’-ATTCTGGTGGTGCTTGTTGTAG tlr5a-qP2-Rev 5’-ACGAGGTAACTTCTGTTCTCAATG tlr5b-qP3-Fw AY389450 5’-GCGTTGTTGAAGAGGCTGGAC tlr5b-qP4-Rev 5’-TTCTGGATGGCCACTTCTCATATTGG mmp9-qP3-Fw NM_213123 5’-CATTAAAGATGCCCTGATGTATCCC mmp9-qP4-Rev 5’-AGTGGTGGTCCGTGGTTGAG il1b-qP1-Fw NM_212844 5’-GAACAGAATGAAGCACATCAAACC il1b-qP2-Rev 5’-ACGGCACTGAATCCACCAC il8-qP1-Fw XM_001342570 5’-TGTGTTATTGTTTTCCTGGCATTTC il8-qP2-Rev 5’-GCGACAGCGTGGATCTACAG ifn1-qP3-Fw NM_207640 5’- TTAATACACGCAAAGATGAGAACTC ifn1-qP4-Rev 5’- GCCAAGCCATTCGCAAGTAG tnfa-qP5-Fw NM_212829 5’- AGACCTTAGACTGGAGAGATGAC tnfa-qP6-Rev 5’- CAAAGACACCTGGCTGTAGAC cxcl-C1c-qP1-Fw NM_001115060 5’- GGCATTCACACCCAAAGCG cxcl-C1c-qP2_Rev 5’- GCGAGCACGATTCACGAGAG * In situ ccl-C5a-Fw NM_001082906 5’- CATCACTAGGAAAGGATTGAAC ccl-C5a-Rev-T7 5’- TAATACGACTCACTATAGGGGATGTCAAAGACTTTATTCAC cxcl-C1c-Fw NM_001115060 5’- GTTAAACATAAATAACACCGACTC cxcl-C1c-Rev-T7 5’- TAATACGACTCACTATAGGGACACCCTATAAAACTGAGTA irak3-Fw CK026195 5’- CAGTGAGAGAGGCATGAAACATC -

Thesis.Pdf (8.850Mb)
Faculty of Biosciences, Fisheries and Economics Photoperiodic history-dependent preadaptation of the smolting gill Novel players and SW immediate response as markers of growth and welfare Marianne Iversen A dissertation for the degree of Philosophiae Doctor, December 2020 Photoperiodic history-dependent preadaptation of the smolting gill Novel players and SW immediate response as markers of growth and welfare Marianne Iversen A dissertation for the degree of Philosophia Doctor December 2020 UiT - The Arctic University of Norway Faculty of Biosciences, Fisheries and Economics Department of Arctic and Marine Biology Arctic Chronobiology and Physiology Research Group Front page image: Juvenile salmon showing the transition from parr (bottom) to smolt (top). Photo by Barbara Tomotani. List of contents I. Acknowledgements ......................................................................................................................... I II. Thesis Abstract ................................................................................................................................ II III. List of papers.................................................................................................................................. III IV. Abbreviations ................................................................................................................................. IV 1. Introduction .................................................................................................................................... 1 -

Genetic Basis of Innovative Anal Fin Pigmentation Patterns in Cichlid Fish
Genetic basis of innovative anal fin pigmentation patterns in cichlid fish Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie Vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Langyu Gu von China Basel, 2016 Originaldokument gespeichert auf dem Dokumentenserver der Universität Basel edoc.unibas.ch Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. Walter Salzburger, Prof. Dr. Alistair McGregor Basel, June, 21st, 2016 Prof. Dr. Jörg Schibler The Dean of Faculty Table of Contents Abstract 1 Chapter 1: Introduction 3 Aim of my PhD project 10 Thesis outline 10 References 11 Chapter 2 Comparative transcriptomics of anal fin pigmentation patterns in cichlid fishes 15 BMC Genomics, 2016, 17: 712. Chapter 3 The genetic basis of convergent evolution of two innovative anal fin pigmentation patterns in East African cichlid fish ——haplochromine eggspots and ectodine blotches 35 Abstract 37 Introduction 38 Materials and Methods 41 Results and Discussion 44 Conclusion 62 Acknowledgements 63 References 63 Supplementary information 73 Chapter 4 Gene network rewiring of the repeated evolution of innovative anal fin pigmentation patterns in cichlid fish 95 Abstract 97 Introduction 98 Methods and Materials 102 Results 104 Discussion 108 Conclusion 111 Acknowledgements 112 References 112 Supplementary information 117 Chapter 5 Expansion via duplication of a multiple-ligand transporter related gene family in cluster in teleost fish 147 Abstract 149 Introduction 149 Materials and Methods 152 Results 156 Discussion 163 Conclusion 168 Acknowledgements 169 References 169 Supplementary files 175 Chapter 6 Discussion and further perspectives 189 References 194 Acknowledgements 197 Curriculum Vitae 199 Abstract The origination of novelty is one of the most fascinating questions in evolutionary biology. -
Androgen Receptor
DOXORUBICIN HYDROCHLORIDE NICLOSAMIDE TNF-alpha Cellular tumor antigen p53Tubulin FLUOROURACIL DAUNORUBICIN HYDROCHLORIDE Hypoxia-inducible factor 1 alpha PACLITAXEL Tubulin beta chain Tubulin alpha chain TOPOTECAN HYDROCHLORIDE Muscleblind-like protein 1 COLCHICINE DIGOXIN Beta-lactamase AmpCFlap endonuclease 1 Ataxin-2 Paired box protein Pax-8 ORF 73 Anthrax lethal factor DIPHENADIONE 6-phospho-1-fructokinasePODOFILOXVINBLASTINE SULFATE Isocitrate dehydrogenase [NADP] cytoplasmicProtein S100-A4 Huntingtin Signal transducer and activator of transcription 6 DNAThioredoxin polymerase reductase beta14-3-3 1- cytoplasmic protein gamma HSP40- subfamily A- putative Menin/Histone-lysineCruzipain N-methyltransferaseNuclear MLL receptor VINCRISTINEsubfamily 0 group SULFATE B NIFENAZONEmember 1 Sodium/potassium-transporting ATPase AZELASTINEMIDAZOLAM HYDROCHLORIDE HYDROCHLORIDE MITOXANTRONE DIHYDROCHLORIDE Solute carrier organic anion transporter Endonucleasefamily memberBISACODYL 4 4C1 Caspase-1 ATPase family AAA domain-containing protein 5 BreastMETHOTREXATE cancer type 1 susceptibility protein TRIFLURIDINE Sodium/potassium-transporting ATPase alpha-1 chain TRIAMTERENECYTARABINE Transcription factor SKN7 AZACITIDINEDEQUALINIUMTRIFLUPROMAZINE CHLORIDEMothersCLADRIBINE against decapentaplegicEPIRUBICIN homolog HYDROCHLORIDE 3 Pteridine reductase 1 Latent membrane protein 1 MELATONIN Arachidonate 15-lipoxygenase- type II Nuclear receptor coactivator 3 DIGITOXIN Retinoid X receptor alpha AMSACRINE IRINOTECAN HYDROCHLORIDE HYDRATE MEBENDAZOLEBacterial -

The Effect of Clozapine on Mrna Expression for Genes Encoding G Protein-Coupled Receptors and the Protein Components of Clathrin-Mediated Endocytosis Sally I
Original article 153 The effect of clozapine on mRNA expression for genes encoding G protein-coupled receptors and the protein components of clathrin-mediated endocytosis Sally I. Sharpa, Ying Huc, Jon F. Weymera, Mie Riziga, Andrew McQuillina, Stephen P. Huntb and Hugh M.D. Gurlinga Objectives Clathrin-mediated endocytosis (CME) is an cells incubated with 5 lmol/l clozapine for 24 h. EDG4 intracellular trafficking mechanism for packaging cargo, expression was also increased with 10–20 lmol/l including G protein-coupled receptors (GPCRs), into clozapine treatment at 48–72 h. Clozapine significantly clathrin-coated vesicles (CCVs). The antipsychotic decreased the expression of b-arrestin, involved in GPCR chlorpromazine inhibits CCV assembly of adaptor protein desensitization, both in vitro and vivo. AP2 whereas clozapine increases serotonin2A receptor Conclusion The changes we report in CME and GPCR internalization. We hypothesized that clozapine alters the mRNAs implicate CCV-mediated internalization of GPCRs expression of CME genes modulating vesicle turnover and the serotonergic system in clozapine’s mechanism of and GPCR internalization. action, which may be useful in the design of more effective Materials and methods SH-SY5Y human neuroblastoma and less toxic antipsychotic therapies. Psychiatr Genet cells were incubated with clozapine (1–20 lmol/l) for 23:153–162 c 2013 Wolters Kluwer Health | Lippincott 24–72 h. GPCR and CME-related gene mRNA expression Williams & Wilkins. was measured using RT-PCR. We quantified changes in the Psychiatric Genetics 2013, 23:153–162 same genes using expression data from a microarray study of mice brains after 12 weeks of treatment with 12 mg/kg/ Keywords: clathrin-coated vesicles, clathrin-mediated endocytosis, clozapine, schizophrenia day clozapine. -

A Fluorescent Sensor for the Real-Time Measurement of Extracellular Oxytocin Dynamics in the Brain
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454450; this version posted July 31, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 2 A fluorescent sensor for the real-time measurement of extracellular 3 oxytocin dynamics in the brain 4 5 Daisuke Ino1, 2*, Hiroshi Hibino2 and Masaaki Nishiyama1 6 7 1. Department of Histology and Cell Biology, Graduate School of Medical Sciences, 8 Kanazawa University, Japan 9 2. Department of Pharmacology, Graduate School of Medicine, Osaka University, Japan 10 11 12 *To whom correspondence should be addressed: 2-2 Yamada-oka, Suita, Osaka 565-0871, 13 Japan 14 Tel: +81-6-6879-3512 15 Fax: +81-6-6879-3519 16 E-mail: [email protected] 17 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454450; this version posted July 31, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 18 ABSTRACT 19 Oxytocin (OT), a hypothalamic neuropeptide that acts as a neuromodulator in the brain, 20 orchestrates a variety of animal behaviors. However, the relationship between brain OT 21 dynamics and complex animal behaviors remains largely elusive, partly because of the lack of 22 a suitable technique for its real-time recording in vivo. Here, we describe MTRIAOT, a G 23 protein-coupled receptor-based green fluorescent OT sensor with a large dynamic range, 24 optimal affinity, ligand specificity to OT orthologs, minimal effects on downstream signaling, 25 and long-term fluorescence stability. -
Investigation of Genetic and Molecular Susceptibility Factors for Bipolar Disorder
Investigation of genetic and molecular susceptibility factors for bipolar disorder Cristiana Cruceanu Department of Human Genetics Faculty of Medicine McGill University, Montreal, Canada March 2016 A thesis submitted to McGill University in partial fulfillment of the requirements of the degree of Doctor of Philosophy © Copyright Cristiana Cruceanu 2016 1 Table of contents TABLE OF CONTENTS --------------------------------------------------------------------------------------------------------------------------------- 2 ABSTRACT/RESUME ---------------------------------------------------------------------------------------------------------------------------------- 3 ACKNOWLEDGEMENTS ------------------------------------------------------------------------------------------------------------------------------ 5 LIST OF ABBREVIATIONS----------------------------------------------------------------------------------------------------------------------------- 7 LIST OF FIGURES ------------------------------------------------------------------------------------------------------------------------------------- 12 LIST OF TABLES --------------------------------------------------------------------------------------------------------------------------------------- 13 PREFACE ------------------------------------------------------------------------------------------------------------------------------------------------ 14 CONTRIBUTION OF AUTHORS ----------------------------------------------------------------------------------------------------------------------------------------------