Unix at the Command Line for More Information? Exercises
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CS101 Lecture 9
How do you copy/move/rename/remove files? How do you create a directory ? What is redirection and piping? Readings: See CCSO’s Unix pages and 9-2 cp option file1 file2 First Version cp file1 file2 file3 … dirname Second Version This is one version of the cp command. file2 is created and the contents of file1 are copied into file2. If file2 already exits, it This version copies the files file1, file2, file3,… into the directory will be replaced with a new one. dirname. where option is -i Protects you from overwriting an existing file by asking you for a yes or no before it copies a file with an existing name. -r Can be used to copy directories and all their contents into a new directory 9-3 9-4 cs101 jsmith cs101 jsmith pwd data data mp1 pwd mp1 {FILES: mp1_data.m, mp1.m } {FILES: mp1_data.m, mp1.m } Copy the file named mp1_data.m from the cs101/data Copy the file named mp1_data.m from the cs101/data directory into the pwd. directory into the mp1 directory. > cp ~cs101/data/mp1_data.m . > cp ~cs101/data/mp1_data.m mp1 The (.) dot means “here”, that is, your pwd. 9-5 The (.) dot means “here”, that is, your pwd. 9-6 Example: To create a new directory named “temp” and to copy mv option file1 file2 First Version the contents of an existing directory named mp1 into temp, This is one version of the mv command. file1 is renamed file2. where option is -i Protects you from overwriting an existing file by asking you > cp -r mp1 temp for a yes or no before it copies a file with an existing name. -

State Notation Language and Sequencer Users' Guide
State Notation Language and Sequencer Users’ Guide Release 2.0.99 William Lupton ([email protected]) Benjamin Franksen ([email protected]) June 16, 2010 CONTENTS 1 Introduction 3 1.1 About.................................................3 1.2 Acknowledgements.........................................3 1.3 Copyright...............................................3 1.4 Note on Versions...........................................4 1.5 Notes on Release 2.1.........................................4 1.6 Notes on Releases 2.0.0 to 2.0.12..................................7 1.7 Notes on Release 2.0.........................................9 1.8 Notes on Release 1.9......................................... 12 2 Installation 13 2.1 Prerequisites............................................. 13 2.2 Download............................................... 13 2.3 Unpack................................................ 14 2.4 Configure and Build......................................... 14 2.5 Building the Manual......................................... 14 2.6 Test.................................................. 15 2.7 Use.................................................. 16 2.8 Report Bugs............................................. 16 2.9 Contribute.............................................. 16 3 Tutorial 19 3.1 The State Transition Diagram.................................... 19 3.2 Elements of the State Notation Language.............................. 19 3.3 A Complete State Program...................................... 20 3.4 Adding a -

Lab Exercise – Ipv4
Lab Exercise – IPv4 Objective To learn about the details of IP (Internet Protocol). IP is the network layer protocol used throughout the Internet. We will examine IP version 4, since it is ubiquitously deployed, while the IP version 6 is partly deployed. The trace is here: https://kevincurran.org/com320/labs/wireshark/trace-ipv4.pcap The text file is here: https://kevincurran.org/com320/labs/wireshark/trace-ipv4.txt Requirements Wireshark: This lab uses the Wireshark software tool to capture and examine a packet trace. A packet trace is a record of traffic at a location on the network, as if a snapshot was taken of all the bits that passed across a particular wire. The packet trace records a timestamp for each packet, along with the bits that make up the packet, from the lower-layer headers to the higher-layer contents. Wireshark runs on most operating systems, including Windows, Mac and Linux. It provides a graphical UI that shows the sequence of packets and the meaning of the bits when interpreted as protocol headers and data. It col- or-codes packets by their type, and has various ways to filter and analyze packets to let you investigate the behavior of network protocols. Wireshark is widely used to troubleshoot networks. You can down- load it from www.wireshark.org if it is not already installed on your computer. We highly recommend that you watch the short, 5 minute video “Introduction to Wireshark” that is on the site. wget / curl: This lab uses wget (Linux and Windows) and curl (Mac) to fetch web resources. -
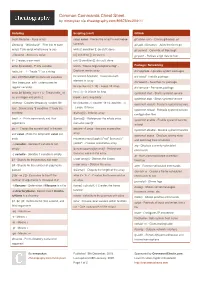
Common Commands Cheat Sheet by Mmorykan Via Cheatography.Com/89673/Cs/20411
Common Commands Cheat Sheet by mmorykan via cheatography.com/89673/cs/20411/ Scripting Scripting (cont) GitHub bash filename - Runs script sleep value - Forces the script to wait value git clone <url > - Clones gitkeeper url Shebang - "# !bi n/b ash " - First line of bash seconds git add <fil ena me> - Adds the file to git script. Tells script what binary to use while [[ condition ]]; do stuff; done git commit - Commits all files to git ./file name - Also runs script if [[ condition ]]; do stuff; fi git push - Pushes all git files to host # - Creates a comment until [[ condition ]]; do stuff; done echo ${varia ble} - Prints variable words=" h ouse dogs telephone dog" - Package / Networking hello_int = 1 - Treats "1 " as a string Declares words array dnf upgrade - Updates system packages Use UPPERC ASE for constant variables for word in ${words} - traverses each dnf install - Installs package element in array Use lowerc ase _wi th_ und ers cores for dnf search - Searches for package for counter in {1..10} - Loops 10 times regular variables dnf remove - Removes package for ((;;)) - Is infinite for loop echo $(( ${hello _int} + 1 )) - Treats hello_int systemctl start - Starts systemd service as an integer and prints 2 break - exits loop body systemctl stop - Stops systemd service mktemp - Creates temporary random file for ((count er=1; counter -le 10; counter ++)) systemctl restart - Restarts systemd service test - Denoted by "[[ condition ]]" tests the - Loops 10 times systemctl reload - Reloads systemd service condition -

A Rapid, Sensitive, Scalable Method for Precision Run-On Sequencing
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.18.102277; this version posted May 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 A rapid, sensitive, scalable method for 2 Precision Run-On sequencing (PRO-seq) 3 4 Julius Judd1*, Luke A. Wojenski2*, Lauren M. Wainman2*, Nathaniel D. Tippens1, Edward J. 5 Rice3, Alexis Dziubek1,2, Geno J. Villafano2, Erin M. Wissink1, Philip Versluis1, Lina Bagepalli1, 6 Sagar R. Shah1, Dig B. Mahat1#a, Jacob M. Tome1#b, Charles G. Danko3,4, John T. Lis1†, 7 Leighton J. Core2† 8 9 *Equal contribution 10 1Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA 11 2Department of Molecular and Cell Biology, Institute of Systems Genomics, University of 12 Connecticut, Storrs, CT 06269, USA 13 3Baker Institute for Animal Health, College of Veterinary Medicine, Cornell University, Ithaca, 14 NY 14853, USA 15 4Department of Biomedical Sciences, College of Veterinary Medicine, Cornell University, 16 Ithaca, NY 14853, USA 17 #aPresent address: Koch Institute for Integrative Cancer Research, Massachusetts Institute of 18 Technology, Cambridge, MA 02139, USA 19 #bPresent address: Department of Genome Sciences, University of Washington, Seattle, WA, 20 USA 21 †Correspondence to: [email protected] or [email protected] 22 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.18.102277; this version posted May 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

K30503041: Creating a Virtual Server with ARP And/Or ICMP Disabled at Creation
K30503041: Creating a virtual server with ARP and/or ICMP disabled at creation Non-Diagnostic Original Publication Date: Apr 16, 2020 Update Date: Apr 16, 2020 Topic You should consider using this procedure under the following condition: You want to create a new virtual server with the Address Resolution Protocol (ARP) and/or the Internet Control Message Protocol (ICMP) disabled at creation. Description In some network environments, you may want to create virtual servers that have ARP and/or ICMP disabled. When creating a new virtual server (one that does not use an IP address of an existing virtual server), you can enable or disable the virtual server. However, you cannot set the IP address of the virtual server with ARP and/or ICMP to 'disabled' when creating the virtual server. You can only enable or disable ARP or ICMP by modifying the virtual address that is automatically created when you create the virtual server. In this scenario, you create the virtual address with ARP and ICMP enabled at the time of creation. You can work around this scenario by creating the virtual address for the virtual server in advance with ARP and/or ICMP disabled. After creating the virtual address, you can then create the virtual server that references the same IP address of the virtual address as the destination IP address of the virtual server. Prerequisites You must meet the following prerequisite to use this procedure: Administrative access to the BIG-IP Configuration utility or the TMOS Shell (tmsh). Note: You cannot create a virtual address using the Configuration utility. -

Openssh-Ldap-Pubkey Documentation Release 0.3.0
openssh-ldap-pubkey Documentation Release 0.3.0 Kouhei Maeda May 18, 2020 Contents 1 openssh-ldap-pubkey 3 1.1 Status...................................................3 1.2 Requirements...............................................3 1.3 See also..................................................3 2 How to setup LDAP server for openssh-lpk5 2.1 Precondition...............................................5 2.2 Requirements...............................................5 2.3 Install...................................................5 3 How to setup OpenSSH server9 3.1 Precondition...............................................9 3.2 Requirements...............................................9 3.3 Install with nslcd (recommend).....................................9 3.4 Install without nslcd........................................... 11 4 History 13 4.1 0.3.0 (2020-05-18)............................................ 13 4.2 0.2.0 (2018-09-30)............................................ 13 4.3 0.1.3 (2018-08-18)............................................ 13 4.4 0.1.2 (2017-11-25)............................................ 13 4.5 0.1.1 (2015-10-16)............................................ 14 4.6 0.1.0 (2015-10-16)............................................ 14 5 Contributors 15 6 Indices and tables 17 i ii openssh-ldap-pubkey Documentation, Release 0.3.0 Contents: Contents 1 openssh-ldap-pubkey Documentation, Release 0.3.0 2 Contents CHAPTER 1 openssh-ldap-pubkey 1.1 Status 1.2 Requirements 1.2.1 LDAP server • Add openssh-lpk schema. • Add an objectClass ldapPublicKey to user entry. • Add one or more sshPublicKey attribute to user entry. 1.2.2 OpenSSH server • OpenSSH over 6.2. • Installing this utility. • Setup AuthorozedKeysCommand and AuthorizedKeysCommandUser in sshd_config. 1.3 See also • OpenSSH 6.2 release 3 openssh-ldap-pubkey Documentation, Release 0.3.0 • openssh-lpk 4 Chapter 1. openssh-ldap-pubkey CHAPTER 2 How to setup LDAP server for openssh-lpk 2.1 Precondition This article restricts OpenLDAP with slapd_config on Debian systems only. -

Specification of the "Sequ" Command
1 Specification of the "sequ" command 2 Copyright © 2013 Bart Massey 3 Revision 0, 1 October 2013 4 This specification describes the "universal sequence" command sequ. The sequ command is a 5 backward-compatible set of extensions to the UNIX [seq] 6 (http://www.gnu.org/software/coreutils/manual/html_node/seq-invo 7 cation.html) command. There are many implementations of seq out there: this 8 specification is built on the seq supplied with GNU Coreutils version 8.21. 9 The seq command emits a monotonically increasing sequence of numbers. It is most commonly 10 used in shell scripting: 11 TOTAL=0 12 for i in `seq 1 10` 13 do 14 TOTAL=`expr $i + $TOTAL` 15 done 16 echo $TOTAL 17 prints 55 on standard output. The full sequ command does this basic counting operation, plus 18 much more. 19 This specification of sequ is in several stages, known as compliance levels. Each compliance 20 level adds required functionality to the sequ specification. Level 1 compliance is equivalent to 21 the Coreutils seq command. 22 The usual specification language applies to this document: MAY, SHOULD, MUST (and their 23 negations) are used in the standard fashion. 24 Wherever the specification indicates an error, a conforming sequ implementation MUST 25 immediately issue appropriate error message specific to the problem. The implementation then 26 MUST exit, with a status indicating failure to the invoking process or system. On UNIX systems, 27 the error MUST be indicated by exiting with status code 1. 28 When a conforming sequ implementation successfully completes its output, it MUST 29 immediately exit, with a status indicating success to the invoking process or systems. -

Curl User Survey 2019 Analysis
curl user survey 2019 analysis “there are so many useful features” summary and analysis by Daniel Stenberg version 1 - June 4, 2019 Table of Contents About curl.............................................................................................................................................3 Survey Background..........................................................................................................................3 Number of responses............................................................................................................................4 Returning respondents?........................................................................................................................5 Users living where?..............................................................................................................................6 What kind of users?..............................................................................................................................7 What protocols......................................................................................................................................8 Multiple platforms..............................................................................................................................10 What platforms...................................................................................................................................11 Which Windows versions...................................................................................................................13 -

Institutionen För Datavetenskap Department of Computer and Information Science
Institutionen för datavetenskap Department of Computer and Information Science Final thesis NetworkPerf – A tool for the investigation of TCP/IP network performance at Saab Transpondertech by Magnus Johansson LIU-IDA/LITH-EX-A--09/039--SE 2009-08-13 Linköpings universitet Linköpings universitet SE-581 83 Linköping, Sweden 581 83 Linköping Final thesis NetworkPerf - A tool for the investigation of TCP/IP network performance at Saab Transpondertech Version 1.0.2 by Magnus Johansson LIU-IDA/LITH-EX-A09/039SE 2009-08-13 Supervisor: Hannes Persson, Attentec AB Examiner: Dr Juha Takkinen, IDA, Linköpings universitet Abstract In order to detect network changes and network troubles, Saab Transpon- dertech needs a tool that can make network measurements. The purpose of this thesis has been to nd measurable network proper- ties that best reect the status of a network, to nd methods to measure these properties and to implement these methods in one single tool. The resulting tool is called NetworkPerf and can measure the following network properties: availability, round-trip delay, delay variation, number of hops, intermediate hosts, available bandwidth, available ports, and maximum al- lowed packet size. The thesis also presents the methods used for measuring these properties in the tool: ping, traceroute, port scanning, and bandwidth measurement. iii iv Acknowledgments This master's thesis would not be half as good as it is, if I had not received help and support from several people. Many thanks to my examiner Dr Juha Takkinen, without whose countin- uous feedback this report would not have been more than a few confusing pages. -

Unix (And Linux)
AWK....................................................................................................................................4 BC .....................................................................................................................................11 CHGRP .............................................................................................................................16 CHMOD.............................................................................................................................19 CHOWN ............................................................................................................................26 CP .....................................................................................................................................29 CRON................................................................................................................................34 CSH...................................................................................................................................36 CUT...................................................................................................................................71 DATE ................................................................................................................................75 DF .....................................................................................................................................79 DIFF ..................................................................................................................................84 -
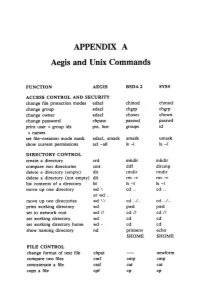
APPENDIX a Aegis and Unix Commands
APPENDIX A Aegis and Unix Commands FUNCTION AEGIS BSD4.2 SYSS ACCESS CONTROL AND SECURITY change file protection modes edacl chmod chmod change group edacl chgrp chgrp change owner edacl chown chown change password chpass passwd passwd print user + group ids pst, lusr groups id +names set file-creation mode mask edacl, umask umask umask show current permissions acl -all Is -I Is -I DIRECTORY CONTROL create a directory crd mkdir mkdir compare two directories cmt diff dircmp delete a directory (empty) dlt rmdir rmdir delete a directory (not empty) dlt rm -r rm -r list contents of a directory ld Is -I Is -I move up one directory wd \ cd .. cd .. or wd .. move up two directories wd \\ cd . ./ .. cd . ./ .. print working directory wd pwd pwd set to network root wd II cd II cd II set working directory wd cd cd set working directory home wd- cd cd show naming directory nd printenv echo $HOME $HOME FILE CONTROL change format of text file chpat newform compare two files emf cmp cmp concatenate a file catf cat cat copy a file cpf cp cp Using and Administering an Apollo Network 265 copy std input to std output tee tee tee + files create a (symbolic) link crl In -s In -s delete a file dlf rm rm maintain an archive a ref ar ar move a file mvf mv mv dump a file dmpf od od print checksum and block- salvol -a sum sum -count of file rename a file chn mv mv search a file for a pattern fpat grep grep search or reject lines cmsrf comm comm common to 2 sorted files translate characters tic tr tr SHELL SCRIPT TOOLS condition evaluation tools existf test test