Dr. Paul Hebert CV
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Forestry Department Food and Agriculture Organization of the United Nations
Forestry Department Food and Agriculture Organization of the United Nations Forest Health & Biosecurity Working Papers OVERVIEW OF FOREST PESTS ROMANIA January 2007 Forest Resources Development Service Working Paper FBS/28E Forest Management Division FAO, Rome, Italy Forestry Department DISCLAIMER The aim of this document is to give an overview of the forest pest1 situation in Romania. It is not intended to be a comprehensive review. The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. © FAO 2007 1 Pest: Any species, strain or biotype of plant, animal or pathogenic agent injurious to plants or plant products (FAO, 2004). Overview of forest pests - Romania TABLE OF CONTENTS Introduction..................................................................................................................... 1 Forest pests and diseases................................................................................................. 1 Naturally regenerating forests..................................................................................... 1 Insects ..................................................................................................................... 1 Diseases................................................................................................................ -

Kobe University Repository : Kernel
Kobe University Repository : Kernel タイトル Revision of braconine wasps of Japan (Hymenoptera: Braconidae) with Title revised generic records 著者 KITTEL, N. Rebecca / QUICKE, L.J. Donald / MAETO, Kaoru Author(s) 掲載誌・巻号・ページ Japanese Journal of Systematic Entomology,25(2):132–153 Citation 刊行日 2019-12-30 Issue date 資源タイプ Journal Article / 学術雑誌論文 Resource Type 版区分 publisher Resource Version 権利 Rights DOI JaLCDOI URL http://www.lib.kobe-u.ac.jp/handle_kernel/90007850 PDF issue: 2021-09-30 Japanese Journal of Systematic Entomology, 25 (2): 132–153. December 30, 2019. Revision of braconine wasps of Japan (Hymenoptera: Braconidae) with revised generic records Rebecca N. KITTEL1), Donald L.J. QUICKE2), and Kaoru MAETO1) 1) Laboratory of Insect Biodiversity and Ecosystem Science, Graduate School of Agricultural Science, Kobe University, Rokkodai 1-1, Nada, Kobe, 657-8501, Japan 2) Department of Biology, Faculty of Science, Chulalongkorn University, Phayathai Road, Bangkok 10330, Thailand E-mail: [email protected] (RNK) / [email protected] (DLJQ) / [email protected] (KM) Abstract The braconine fauna of Japan is revised, based on literature and on the collections of the Osaka Museum of Natural History, Osaka, and the Institute for Agro-Environmental Sciences, Tsukuba. A key to the genera is included and distribution records are provided at the prefecture level. Two genera (Baryproctus Ashmead and Dioxybracon Granger) are recorded for the first time from Japan, with the species Baryproctus barypus (Marshall) and Dioxybracon koshunensis (Watanabe) comb. nov. (= Bracon koshunensis Watanabe). The two species Stenobracon oculatus and Chelonogastra formosana are excluded from the Japanese species list. -
American Horticulturist Volume 72, Number 2 February 1993
American Horticulturist Volume 72, Number 2 February 1993 ARTICLES Proven Performers In our popular annual feature, three national plant societies name some (nearly) fail-safe favorites. Dianthuses by Rand B. Lee ......................................... 12 African Violets by Carol Bruce ......................................... 17 Lilies by Calvin Helsley .. .......................... .......... 21 Men Who've Loved Lilies by Melissa Dodd Eskilson .................. ...... ....... 26 From the exquisite but fussy species, lily-breeding pioneers have produced tough-as-nails hybrids for gardeners and florists. FEBRUARY'S COVER Drip Rationale Photographed by Priscilla Eastman by Robert Kourik ....................................... 34 The three-foot-tall Vollmer's tiger Simple hardware offers a drought-busting, water-conserving path lily, Lilium vollmeri, grows in to lusher growth. hillside bogs in two counties in southwest Oregon and adjacent A Defense of Ailanthus areas of California. It is threatened by Richard S. Peigler .... .. ... ......................... 38 by collecting throughout its range, according to Donald C. Eastman's It may be the stinking ash to some, but in a city lot bereft of other Rare and Endangered Plants of greenery, it earns the name tree-of-heaven. Oregon. Of ninety lily species native to the Northern hemisphere, only twenty-two have been tapped by breeders for garden and DEPARTMENTS cut-flower hybrids. The Nature Conservancy reports that at least Commentary .. ... .... .. ............. ... ... ............ 4 seven -

Wildlife Matters: Winter 2016 1 Wildlife Matters
Wildlife Matters: Winter 2016 1 wildlife matters Winter 2016 Historic partnership: AWC to reintroduce lost mammals to NSW national parks 2 Wildlife Matters: Winter 2016 Saving Australia’s threatened wildlife Welcome to the Winter 2016 edition of Wildlife Matters. The AWC mission This edition marks the beginning of a historic partnership between Australian The mission of Australian Wildlife Wildlife Conservancy (AWC) and the NSW Government. AWC has been contracted Conservancy (AWC) is the effective to deliver national park management services in the iconic Pilliga forest and at conservation of all Australian animal Mallee Cliffs National Park in the state’s south-west. It is the first public-private species and the habitats in which they live. collaboration of its kind. The centrepiece of this exciting partnership will be the reintroduction of at least 10 mammal species that are currently listed as extinct in To achieve this mission our actions are NSW. focused on: This is one of the world’s most significant biodiversity reconstruction projects. The • Establishing a network of sanctuaries return of mammals such as the Bilby and the Numbat – which disappeared from which protect threatened wildlife and NSW national parks more than 100 years ago – will represent a defining moment in ecosystems: AWC now manages our quest to halt and reverse the loss of Australia’s unique wildlife. 25 sanctuaries covering over 3.25 million hectares (8 million acres). The initiative reflects strong leadership by the NSW Government. It is committing substantial funds for threatened species, including this partnership with AWC. • Implementing practical, on-ground More importantly, the NSW Government recognises the need to develop new conservation programs to protect approaches to conservation if we are to reverse the catastrophic decline of the wildlife at our sanctuaries: these Australia’s natural capital. -
A New Species of the Genus Hyalella (Crustacea, Amphipoda) from Northern Mexico
ZooKeys 942: 1–19 (2020) A peer-reviewed open-access journal doi: 10.3897/zookeys.942.50399 RESEARCH ARTicLE https://zookeys.pensoft.net Launched to accelerate biodiversity research A new species of the genus Hyalella (Crustacea, Amphipoda) from northern Mexico Aurora Marrón-Becerra1, Margarita Hermoso-Salazar2, Gerardo Rivas2 1 Posgrado en Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México; Av. Ciudad Univer- sitaria 3000, C.P. 04510, Coyoacán, Ciudad de México, México 2 Facultad de Ciencias, Universidad Nacional Autónoma de México; Av. Ciudad Universitaria 3000, C.P. 04510, Coyoacán, Ciudad de México, México Corresponding author: Aurora Marrón-Becerra ([email protected]) Academic editor: T. Horton | Received 22 January 2020 | Accepted 4 May 2020 | Published 18 June 2020 http://zoobank.org/85822F2E-D873-4CE3-AFFB-A10E85D7539F Citation: Marrón-Becerra A, Hermoso-Salazar M, Rivas G (2020) A new species of the genus Hyalella (Crustacea, Amphipoda) from northern Mexico. ZooKeys 942: 1–19. https://doi.org/10.3897/zookeys.942.50399 Abstract A new species, Hyalella tepehuana sp. nov., is described from Durango state, Mexico, a region where stud- ies on Hyalella have been few. This species differs from most species of the North and South American genus Hyalella in the number of setae on the inner plate of maxilla 1 and maxilla 2, characters it shares with Hyalella faxoni Stebbing, 1903. Nevertheless, H. faxoni, from the Volcan Barva in Costa Rica, lacks a dorsal process on pereionites 1 and 2. Also, this new species differs from other described Hyalella species in Mex- ico by the shape of the palp on maxilla 1, the number of setae on the uropods, and the shape of the telson. -

Lepidoptera Collecting in Kenya and Tanzania
Vol. 4 No. 1 1993 BROS: Kenya and Tanzania Lepidoptera 17 TROPICAL LEPIDOPTERA, 4(1): 16-25 LEPIDOPTERA COLLECTING IN KENYA AND TANZANIA EMMANUEL BROS DE PUECHREDON1 "La Fleurie," Rebgasse 28, CH-4102 Binningen BL, Switzerland ABSTRACT.- Situated in tropical Africa, on both sides of the Equator, Kenya and Tanzania possess an extraordinary rich Lepidoptera fauna (according to Larsen's latest book on Kenya: 871 species only for the Rhopalocera and Grypocera). The present paper reports on the author's participation in a non-entomological mini-expedition during January 1977 across those two countries, with comments on the areas where collecting was possible and practiced by him as a serious amateur lepidopterist. In addition there are photos of some interesting landscapes and, last but not least, a complete list of all the species captured and noted. RESUME.- En pleine Afrique equatoriale, a cheval sur 1'Equateur, le Kenya et la Tanzanie possedent une faune de Lepidopteres extraordinairement riche (871 especes seulement pour les Rhopaloceres et Hesperiides du Kenya, selon le tout recent ouvrage de Larsen). La presente note relate une mini-expedition non specifiquement entomologique en Janvier 1977 a travers ces deux pays, avec commmentaires de 1'auteur, lepidopteriste amateur eclaire, sur les lieux ou il a eu la possibilite de collectionner, recit agremente de quelques photos de biotopes interessants et surtout avec la liste complete des especes capturees et notees. KEY WORDS: Acraeinae, Africa, Arctiidae, Cossidae, Danainae, distribution, Ethiopian, Eupterotidae, Hesperiidae, Limacodidae, Lymantriidae, Noctuidae, Notodontidae, Nymphalidae, Papilionidae, Pieridae, Psychidae, Pyralidae, Saturniidae, Satyrinae, Thaumetopoeinae. In January 1977, I had the opportunity of participating in a Mt. -

15 International Symposium on Ostracoda
Berliner paläobiologische Abhandlungen 1-160 6 Berlin 2005 15th International Symposium on Ostracoda In Memory of Friedrich-Franz Helmdach (1935-1994) Freie Universität Berlin September 12-15, 2005 Abstract Volume (edited by Rolf Kohring and Benjamin Sames) 2 ---------------------------------------------------------------------------------------------------------------------------------------- Preface The 15th International Symposium on Ostracoda takes place in Berlin in September 2005, hosted by the Institute of Geological Sciences of the Freie Universität Berlin. This is the second time that the International Symposium on Ostracoda has been held in Germany, following the 5th International Symposium in Hamburg in 1974. The relative importance of Ostracodology - the science that studies Ostracoda - in Germany is further highlighted by well-known names such as G.W. Müller, Klie, Triebel and Helmdach, and others who stand for the long tradition of research on Ostracoda in Germany. During our symposium in Berlin more than 150 participants from 36 countries will meet to discuss all aspects of living and fossil Ostracoda. We hope that the scientific communities working on the biology and palaeontology of Ostracoda will benefit from interesting talks and inspiring discussions - in accordance with the symposium's theme: Ostracodology - linking bio- and geosciences We wish every participant a successful symposium and a pleasant stay in Berlin Berlin, July 27th 2005 Michael Schudack and Steffen Mischke CONTENT Schudack, M. and Mischke, S.: Preface -

Stanley I. Dodson Complete List of Publications
Announcements Resolution of Respect Stanley Ivan Dodson 1944–2009 Stanley was born on 26 July 1944 in Lincoln, Illinois to Ivan Frank Dodson and Dorothy Wanda Dodson. When he was 10 years old his family moved to Grand Junction, Colorado where his father had an opportunity to prospect for uranium. It was there that Stanley learned his love of the West, deserts, mountains, and ponds. After graduating from Grand Junction High School in 1962, Stanley attended Yale University and obtained a Bachelor of Science degree in 1966, conducting research with John Brooks for his undergraduate Honors Thesis. While at Yale he met Virginia Elizabeth Joseph, and they married on 16 January 1967. Stanley and Ginny enrolled in doctoral programs with Tommy Edmondson at the University of Washington, where Stanley received his Doctorate in Zoology in 1970. He spent his entire professional career on the faculty in Zoology at the University of Wisconsin in Madison from 1970 until his retirement in 2008. Stanley’s life was cut short on 23 August 2009 when he died of injuries sustained in an accident while bicycling at the Colorado National Monument near Grand Junction. He is survived by his wife, Ginny, his daughter, Sarah Emily Dodson Wilson, his son-in-law, Ian David Sobaczewski Wilson, his grandchildren, Kate Lorraine Wilson and Henry August Wilson, his mother, and his sisters Dian April Dunn (Frank) and Sarah Annette Martin. He was predeceased by his father. Contributions to ecological science Stanley Dodson made many important contributions to aquatic ecology and to ecological science in general. He began his distinguished research career working with John Brooks during his second year as an undergraduate at Yale. -

2009 Editorial
www.limnology.org Volume 55 - December 2009 Editorial While care is taken to accurately report Some sad news but also good news! information, SILnews is not responsible I y.suppose.b .now.many.of.you.heard.the.tragic. these.meetings. for information and/or advertisements news.about.the.deaths.in.August.’09..of.two. briefly.in.this. published herein and does not endorse, of.our.very.distinguished.limnologists:.Dr.. Newsletter. approve or recommend products, programs or opinions expressed. Stanley.Dodson.(University.of..Wisconsin,. as.I.had.the. USA)..and.Dr..W.John.O’.Brien.(University.of. privilege.of. N..Carolina,.USA)...This.Newsletter.contains. participating. their.obituaries.(taken.with.permission.from. in.both.these. Hydrobiologia.and.Journal of Fundamental meetings..My. In This Issue and Applied Limnology,.respectively),.as.well. impression. as.the.obituaries.of..Dr..Thomas.Nogrady. based.on.a. Obituaries........................................2-6 (Kingston,.Ont..Canada).who.died.in.July. visit.to.Lake. ’09.and.Dr..Roger.Pourriot..(France).who.had. Taihu.. Regional.Limnology.Reports..........7-21 died.in.August.’08..I.convey.on.behalf.of.the. and.talking. SILNews.Letter,.the.SIL.Secretariat.and.on.my. to.scientists,. Reports.from.SIL.. own.behalf.our.heartfelt.condolences.to.the. is.that.lake. Working.Groups.and.. bereaved.families.. restoration. other.Conferences........................22-32 The.good.news.is.that.our.SIL.Working. and.pollution. Ramesh Gulati, December 2009 Announcements...........................32-34 Groups.are.quite.active.and.many.of.them. abatements.are. have.sent.their.brief.research.activity.reports. high.up.on.the.agenda.of.the.Chinese.limnolo- Book.Reviews.. or.of.their.planned.studies...Also,.I.am.happy. -

Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A
Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research Julien Thézé, Carlos Lopez-Vaamonde, Jenny Cory, Elisabeth Herniou To cite this version: Julien Thézé, Carlos Lopez-Vaamonde, Jenny Cory, Elisabeth Herniou. Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research. Viruses, MDPI, 2018, 10 (7), pp.366. 10.3390/v10070366. hal-02140538 HAL Id: hal-02140538 https://hal.archives-ouvertes.fr/hal-02140538 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License viruses Article Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research Julien Thézé 1,2, Carlos Lopez-Vaamonde 1,3 ID , Jenny S. Cory 4 and Elisabeth A. Herniou 1,* ID 1 Institut de Recherche sur la Biologie de l’Insecte, UMR 7261, CNRS—Université de Tours, 37200 Tours, France; [email protected] (J.T.); [email protected] -

Bacterial Associates of Orthezia Urticae, Matsucoccus Pini, And
Protoplasma https://doi.org/10.1007/s00709-019-01377-z ORIGINAL ARTICLE Bacterial associates of Orthezia urticae, Matsucoccus pini, and Steingelia gorodetskia - scale insects of archaeoccoid families Ortheziidae, Matsucoccidae, and Steingeliidae (Hemiptera, Coccomorpha) Katarzyna Michalik1 & Teresa Szklarzewicz1 & Małgorzata Kalandyk-Kołodziejczyk2 & Anna Michalik1 Received: 1 February 2019 /Accepted: 2 April 2019 # The Author(s) 2019 Abstract The biological nature, ultrastructure, distribution, and mode of transmission between generations of the microorganisms associ- ated with three species (Orthezia urticae, Matsucoccus pini, Steingelia gorodetskia) of primitive families (archaeococcoids = Orthezioidea) of scale insects were investigated by means of microscopic and molecular methods. In all the specimens of Orthezia urticae and Matsucoccus pini examined, bacteria Wolbachia were identified. In some examined specimens of O. urticae,apartfromWolbachia,bacteriaSodalis were detected. In Steingelia gorodetskia, the bacteria of the genus Sphingomonas were found. In contrast to most plant sap-sucking hemipterans, the bacterial associates of O. urticae, M. pini, and S. gorodetskia are not harbored in specialized bacteriocytes, but are dispersed in the cells of different organs. Ultrastructural observations have shown that bacteria Wolbachia in O. urticae and M. pini, Sodalis in O. urticae, and Sphingomonas in S. gorodetskia are transovarially transmitted from mother to progeny. Keywords Symbiotic microorganisms . Sphingomonas . Sodalis-like -
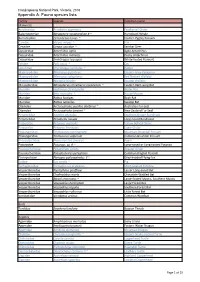
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus