A Description of the Fish Fauna of Uzbekistan Using DNA Barcoding
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Dynamic Genetic Diversity and Population Structure of Coreius Guichenoti
ZooKeys 1055: 135–148 (2021) A peer-reviewed open-access journal doi: 10.3897/zookeys.1055.70117 RESEARCH ARTICLE https://zookeys.pensoft.net Launched to accelerate biodiversity research Dynamic genetic diversity and population structure of Coreius guichenoti Dongqi Liu1*, Feng Lan2*, Sicai Xie1, Yi Diao1, Yi Zheng1, Junhui Gong1 1 Sichuan Province Key Laboratory of Characteristic Biological Resources of Dry and Hot River Valley, Pan- zhihua University, Panzhihua, 617000, China 2 Upper Changjiang River Burean of Hydrological and Water Resources Survey, Chongqing, 400000, China Corresponding author: Feng Lan ([email protected]) Academic editor: M.E. Bichuette | Received 14 June 2021 | Accepted 27 July 2021 | Published 11 August 2021 http://zoobank.org/ADECA19A-B689-47AE-971B-42913F28F5CE Citation: Liu D, Lan F, Xie S, Diao Y, Zheng Y, Gong J (2021) Dynamic genetic diversity and population structure of Coreius guichenoti. ZooKeys 1055: 135–148. https://doi.org/10.3897/zookeys.1055.70117 Abstract To investigate the genetic effects on the population of Coreius guichenoti of dam constructions in the upper reaches of the Yangtze River, we analyzed the genetic diversity and population structure of 12 popula- tions collected in 2009 and 2019 using mitochondrial DNA (mtDNA) control regions. There was no significant difference in genetic diversity between 2009 and 2019P ( > 0.05), but the population structure tended to become stronger. Genetic differentiation (FST) among five populations (LX, BB, YB, SF and JA) collected in 2009 was not significant P( > 0.05). However, some populations collected in 2019 were significantly differentiated (P < 0.05), indicating that the population structure has undergone change. -

Review and Updated Checklist of Freshwater Fishes of Iran: Taxonomy, Distribution and Conservation Status
Iran. J. Ichthyol. (March 2017), 4(Suppl. 1): 1–114 Received: October 18, 2016 © 2017 Iranian Society of Ichthyology Accepted: February 30, 2017 P-ISSN: 2383-1561; E-ISSN: 2383-0964 doi: 10.7508/iji.2017 http://www.ijichthyol.org Review and updated checklist of freshwater fishes of Iran: Taxonomy, distribution and conservation status Hamid Reza ESMAEILI1*, Hamidreza MEHRABAN1, Keivan ABBASI2, Yazdan KEIVANY3, Brian W. COAD4 1Ichthyology and Molecular Systematics Research Laboratory, Zoology Section, Department of Biology, College of Sciences, Shiraz University, Shiraz, Iran 2Inland Waters Aquaculture Research Center. Iranian Fisheries Sciences Research Institute. Agricultural Research, Education and Extension Organization, Bandar Anzali, Iran 3Department of Natural Resources (Fisheries Division), Isfahan University of Technology, Isfahan 84156-83111, Iran 4Canadian Museum of Nature, Ottawa, Ontario, K1P 6P4 Canada *Email: [email protected] Abstract: This checklist aims to reviews and summarize the results of the systematic and zoogeographical research on the Iranian inland ichthyofauna that has been carried out for more than 200 years. Since the work of J.J. Heckel (1846-1849), the number of valid species has increased significantly and the systematic status of many of the species has changed, and reorganization and updating of the published information has become essential. Here we take the opportunity to provide a new and updated checklist of freshwater fishes of Iran based on literature and taxon occurrence data obtained from natural history and new fish collections. This article lists 288 species in 107 genera, 28 families, 22 orders and 3 classes reported from different Iranian basins. However, presence of 23 reported species in Iranian waters needs confirmation by specimens. -

Downloaded and Been Reported (Filonzi, Chiesa, Vaghi, & Nonnis Marzano, 2010; Potential Duplicates Eliminated
Food Control 79 (2017) 297e308 Contents lists available at ScienceDirect Food Control journal homepage: www.elsevier.com/locate/foodcont Novel nuclear barcode regions for the identification of flatfish species Valentina Paracchini a, Mauro Petrillo a, Antoon Lievens a, Antonio Puertas Gallardo a, Jann Thorsten Martinsohn a, Johann Hofherr a, Alain Maquet b, Ana Paula Barbosa Silva b, * Dafni Maria Kagkli a, Maddalena Querci a, Alex Patak a, Alexandre Angers-Loustau a, a European Commission, Joint Research Centre (JRC), via E. Fermi 2749, 21027 Ispra, Italy b European Commission, Joint Research Centre (JRC), Retieseweg 111, 2440 Geel, Belgium article info abstract Article history: The development of an efficient seafood traceability framework is crucial for the management of sus- Received 7 November 2016 tainable fisheries and the monitoring of potential substitution fraud across the food chain. Recent studies Received in revised form have shown the potential of DNA barcoding methods in this framework, with most of the efforts focusing 5 April 2017 on using mitochondrial targets such as the cytochrome oxidase 1 and cytochrome b genes. In this article, Accepted 6 April 2017 we show the identification of novel targets in the nuclear genome, and their associated primers, to be Available online 7 April 2017 used for the efficient identification of flatfishes of the Pleuronectidae family. In addition, different in silico methods are described to generate a dataset of barcode reference sequences from the ever-growing Keywords: Bioinformatics wealth of publicly available sequence information, replacing, where possible, labour-intensive labora- DNA barcoding tory work. The short amplicon lengths render the analysis of these new barcode target regions ideally Next-generation sequencing suited to next-generation sequencing techniques, allowing characterisation of multiple fish species in Seafood identification mixed and processed samples. -

Proof-Of-Concept of Environmental Dna Tools for Atlantic Sturgeon Management
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2015 PROOF-OF-CONCEPT OF ENVIRONMENTAL DNA TOOLS FOR ATLANTIC STURGEON MANAGEMENT Jameson Hinkle Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Animals Commons, Applied Statistics Commons, Biology Commons, Biostatistics Commons, Environmental Health and Protection Commons, Fresh Water Studies Commons, Genetics Commons, Natural Resources and Conservation Commons, Natural Resources Management and Policy Commons, Other Environmental Sciences Commons, Other Genetics and Genomics Commons, Other Life Sciences Commons, and the Water Resource Management Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/3932 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Center for Environmental Studies Virginia Commonwealth University This is to certify that the thesis prepared by Jameson E. Hinkle entitled “ProofofConcept of Environmental DNA tools for Atlantic Sturgeon Management” has been approved by his committee as satisfactory completion of the thesis requirement for the degree of Master of Science in Environmental Studies (M.S. ENVS) _________________________________________________________________________ Greg Garman, Ph.D., Director, Center -
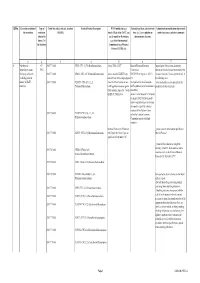
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

Review and Meta-Analysis of the Environmental Biology and Potential Invasiveness of a Poorly-Studied Cyprinid, the Ide Leuciscus Idus
REVIEWS IN FISHERIES SCIENCE & AQUACULTURE https://doi.org/10.1080/23308249.2020.1822280 REVIEW Review and Meta-Analysis of the Environmental Biology and Potential Invasiveness of a Poorly-Studied Cyprinid, the Ide Leuciscus idus Mehis Rohtlaa,b, Lorenzo Vilizzic, Vladimır Kovacd, David Almeidae, Bernice Brewsterf, J. Robert Brittong, Łukasz Głowackic, Michael J. Godardh,i, Ruth Kirkf, Sarah Nienhuisj, Karin H. Olssonh,k, Jan Simonsenl, Michał E. Skora m, Saulius Stakenas_ n, Ali Serhan Tarkanc,o, Nildeniz Topo, Hugo Verreyckenp, Grzegorz ZieRbac, and Gordon H. Coppc,h,q aEstonian Marine Institute, University of Tartu, Tartu, Estonia; bInstitute of Marine Research, Austevoll Research Station, Storebø, Norway; cDepartment of Ecology and Vertebrate Zoology, Faculty of Biology and Environmental Protection, University of Lodz, Łod z, Poland; dDepartment of Ecology, Faculty of Natural Sciences, Comenius University, Bratislava, Slovakia; eDepartment of Basic Medical Sciences, USP-CEU University, Madrid, Spain; fMolecular Parasitology Laboratory, School of Life Sciences, Pharmacy and Chemistry, Kingston University, Kingston-upon-Thames, Surrey, UK; gDepartment of Life and Environmental Sciences, Bournemouth University, Dorset, UK; hCentre for Environment, Fisheries & Aquaculture Science, Lowestoft, Suffolk, UK; iAECOM, Kitchener, Ontario, Canada; jOntario Ministry of Natural Resources and Forestry, Peterborough, Ontario, Canada; kDepartment of Zoology, Tel Aviv University and Inter-University Institute for Marine Sciences in Eilat, Tel Aviv, -

Potential Effects of Dam Cascade on Fish
Rev Fish Biol Fisheries DOI 10.1007/s11160-015-9395-9 ORIGINAL RESEARCH Potential effects of dam cascade on fish: lessons from the Yangtze River Fei Cheng . Wei Li . Leandro Castello . Brian R. Murphy . Songguang Xie Received: 23 October 2014 / Accepted: 13 July 2015 Ó Springer International Publishing Switzerland 2015 Abstract Construction of hydroelectric dams affect Corieus guichenoti will have a high risk of extinction river ecosystems, fish diversity, and fisheries yields. due to the combined effects of impoundment and However, there are no studies assessing the combined blocking. Modification of the flow regime will effects on fish caused by several adjacent dams and adversely affect the recruitment of 26 species that their reservoirs, as in a ‘dam cascade’. This study produce drifting eggs. The start of annual spawning for predicts the potential effects that a cascade of ten dams 13 fishes will be postponed by more than 1 month, and currently under construction in the upper Yangtze fish spawning and growth opportunities will be River in China will have on local fishes, and uses such reduced due to low water temperatures associated predictions to assess the effectiveness of possible fish with hypolimnetic discharges. Combined dam effects conservation measures. We found that the dam will further reduce the likelihood of successful cascade will have serious combined effects on fishes recruitment of some endangered species, such as mainly due to impoundment, habitat fragmentation Acipenser dabryanus and Psephurus gladius. Three and blocking, flow regime modification, and hypolim- countermeasures hold promise to mitigate the near- netic discharges. The impoundments will cause loss of term effects of the dam cascade, including preserva- critical habitats for 46 endemic species. -

Effectiveness of FISK, an Invasiveness Screening Tool for Nonnative
Risk Analysis DOI: 10.1111/risa.12050 Effectiveness of FISK, an Invasiveness Screening Tool for Non-Native Freshwater Fishes, to Perform Risk Identification Assessments in the Iberian Peninsula David Almeida,1,2 Filipe Ribeiro,3,4 Pedro M. Leunda,5,6 Lorenzo Vilizzi,7 and Gordon H. Copp1,2,6,8,∗ Risk assessments are crucial for identifying and mitigating impacts from biological invasions. The Fish Invasiveness Scoring Kit (FISK) is a risk identification (screening) tool for fresh- water fishes consisting of two subject areas: biogeography/history and biology/ecology. Ac- cording to the outcomes, species can be classified under particular risk categories. The aim of this study was to apply FISK to the Iberian Peninsula, a Mediterranean climate region highly important for freshwater fish conservation due to a high level of endemism. In total, 89 fish species were assessed by three independent assessors. Results from receiver operating characteristic analysis showed that FISK can discriminate reliably between noninvasive and invasive fishes for Iberia, with a threshold of 20.25, similar to those obtained in several regions around the world. Based on mean scores, no species was categorized as “low risk,” 50 species as “medium risk,” 17 as “moderately high risk,” 11 as “high risk,” and 11 as “very high risk.” The highest scoring species was goldfish Carassius auratus. Mean certainty in response was above the category “mostly certain,” ranging from tinfoil barb Barbonymus schwanenfeldii with the lowest certainty to eastern mosquitofish Gambusia holbrooki with the highest level. Pair-wise comparison showed significant differences between one assessor and the other two on mean certainty, with these two assessors showing a high coincidence rate for the species categorization. -

Homoplasies, Consistency Index and the Complexity of Morphological Evolution: Catfishes As a Case Study for General Discussions on Phylogeny and Macroevolution
Int. J. Morphol., 25(4):831-837, 2007. Homoplasies, Consistency Index and the Complexity of Morphological Evolution: Catfishes as a Case Study for General Discussions on Phylogeny and Macroevolution Homoplasias, Índice de Consistencia y la Complejidad de la Evolución Morfológica: Peces Gato como un Estudio de Caso para Discusiones Generales en Filogenia y Macroevolución *,** Rui Diogo DIOGO, R. Homoplasies, consistency index and the complexity of morphological evolution: Catfishes as a case study for general discussions on phylogeny and macroevolution. Int. J. Morphol., 25(4):831-837, 2007. SUMMARY: Catfishes constitute a highly diversified, cosmopolitan group that represents about one third of all freshwater fishes and is one of the most diverse Vertebrate taxa. The detailed study of the Siluriformes can, thus, provide useful data, and illustrative examples, for broader discussions on general phylogeny and macroevolution. In this short note I briefly expose how the study of this remarkably diverse group of fishes reveals an example of highly homoplasic, complex 'mosaic' morphological evolution. KEY WORDS: Catfishes; Homoplasies; Morphological macroevolution; Phylogeny; Siluriformes; Teleostei. INTRODUCTION The catfishes, or Siluriformes, found in North, Cen- and diversity surely resulting from several homoplasic tral and South America, Africa, Europe, Asia and Australia, events. This was precisely the main reason to choose this with fossils inclusively found in Antarctica, constitute a amazing group of fishes as a case study for discussing gene- highly diversified, cosmopolitan group, which, with more ral topics on phylogeny and macroevolution. But the exam than 2700 species, represents about one third of all freshwater of more and more morphological phylogenetic characters in fishes and is one of the most diverse Vertebrate taxa (e.g. -

Family-Cyprinidae-Gobioninae-PDF
SUBFAMILY Gobioninae Bleeker, 1863 - gudgeons [=Gobiones, Gobiobotinae, Armatogobionina, Sarcochilichthyna, Pseudogobioninae] GENUS Abbottina Jordan & Fowler, 1903 - gudgeons, abbottinas [=Pseudogobiops] Species Abbottina binhi Nguyen, in Nguyen & Ngo, 2001 - Cao Bang abbottina Species Abbottina liaoningensis Qin, in Lui & Qin et al., 1987 - Yingkou abbottina Species Abbottina obtusirostris (Wu & Wang, 1931) - Chengtu abbottina Species Abbottina rivularis (Basilewsky, 1855) - North Chinese abbottina [=lalinensis, psegma, sinensis] GENUS Acanthogobio Herzenstein, 1892 - gudgeons Species Acanthogobio guentheri Herzenstein, 1892 - Sinin gudgeon GENUS Belligobio Jordan & Hubbs, 1925 - gudgeons [=Hemibarboides] Species Belligobio nummifer (Boulenger, 1901) - Ningpo gudgeon [=tientaiensis] Species Belligobio pengxianensis Luo et al., 1977 - Sichuan gudgeon GENUS Biwia Jordan & Fowler, 1903 - gudgeons, biwas Species Biwia springeri (Banarescu & Nalbant, 1973) - Springer's gudgeon Species Biwia tama Oshima, 1957 - tama gudgeon Species Biwia yodoensis Kawase & Hosoya, 2010 - Yodo gudgeon Species Biwia zezera (Ishikawa, 1895) - Biwa gudgeon GENUS Coreius Jordan & Starks, 1905 - gudgeons [=Coripareius] Species Coreius cetopsis (Kner, 1867) - cetopsis gudgeon Species Coreius guichenoti (Sauvage & Dabry de Thiersant, 1874) - largemouth bronze gudgeon [=platygnathus, zeni] Species Coreius heterodon (Bleeker, 1865) - bronze gudgeon [=rathbuni, styani] Species Coreius septentrionalis (Nichols, 1925) - Chinese bronze gudgeon [=longibarbus] GENUS Coreoleuciscus -

Gobio Gobio (Linnaeus, 1758). Gobio AUTÓCTONA Catalán; Gobi
Atlas y Libro Rojo de los Peces Continentales de España ESPECIE Gobio gobio (Linnaeus, 1758). Gobio AUTÓCTONA Catalán; Gobi. Vasco; Gobioa. Portugués; GÓbio. DESCRIPCIÓN Ciprínido de pequeño tamaño, que raramente supera los 15 cm de longitud. La forma de su cuer po refleja su condición de pez de fondo con una parte ventral más o menos plana, un dorso conve xo y la boca ínfera, con una barbilla a cada lado. El co lor del dorso es oscuro y en los flancos aparece una Clase: Actinopterygii línea de manchas redondeadas y azuladas sobre un Orden: Cypriniformes fondo más claro. La aleta dorsal y aleta caudal, fuerte Familia: Cyprinidae mente escotada, están irregularmente punteadas. Tiene Sinonimias: Cyprinus gobio Linnaeus, 1758. Leuciscus entre 37 y 41 escamas en la línea lateral. gobio (Linnaeus, 1758). Cobítis fundulus Wulff, 1765. Gobio fluviatilís Fleming, 1828. Gobio phoxinoides De BIOLOGÍA Y ECOLOGÍA la Pylaie, 1835. Gobio vulgaris Heckel, 1836. Gobio sa xatilis Koch, 1840. Gobio obtusirostris Valenciennes, Los adultos pueden encontrarse en fondos más o 1842. Gobio fluviatilis Cuvier, 1842. Bungia nigrescens menos blandos o cubiertos por sedimentos y también Keyserling, 1861. Gobio fluviatílís cynocephalus Dy en ambientes más lénticos, siempre que haya lugares bowski, 1869. Gobio fluviatilis lepidolaemus Kessler, cercanos donde existan las condiciones necesarias para 1872. Gobio gobio lepidolaemus Kessler, 1872. Gobio le su reproducción Se alimenta básicamente de macroin pidolaemus Kessler, 1872. Gobio lepidolaemus caucasi ca Kamensky, 1901. Gobio latus Ankin, 1905. Gobio go vertebrados bentónicos (larvas de insectos, crustáceos bio brevicirris Berg, 1914. Gobio gobio katopyga Berg, y moluscos). 1914. Gobio gobio longicirris Berg, 1914. -

SCIENCE CHINA Phylogenetic Relationships and Estimation Of
SCIENCE CHINA Life Sciences • RESEARCH PAPER • April 2012 Vol.55 No.4: 312–320 doi: 10.1007/s11427-012-4305-z Phylogenetic relationships and estimation of divergence times among Sisoridae catfishes YU MeiLing1,2* & HE ShunPing1* 1Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 400732, China; 2Graduate University of Chinese Academy of Sciences, Beijing 100049, China Received December 10, 2011; accepted March 9, 2012 Nineteen taxa representing 10 genera of Sisoridae were subjected to phylogenetic analyses of sequence data for the nuclear genes Plagl2 and ADNP and the mitochondrial gene cytochrome b. The three data sets were analyzed separately and combined into a single data set to reconstruct phylogenetic relationships among Chinese sisorids. Both Chinese Sisoridae as a whole and the glyptosternoid taxa formed monophyletic groups. The genus Pseudecheneis is likely to be the earliest diverging extant ge- nus among the Chinese Sisoridae. The four Pareuchiloglanis species included in the study formed a monophyletic group. Glaridoglanis was indicated to be earliest diverging glyptosternoid, followed by Glyptosternon maculatum and Exostoma labi- atum. Our data supported the conclusion that Oreoglanis and Pseudexostoma both formed a monophyletic group. On the basis of the fossil record and the results of a molecular dating analysis, we estimated that the Sisoridae diverged in the late Miocene about 12.2 Mya. The glyptosternoid clade was indicated to have diverged, also in the late Miocene, about 10.7 Mya, and the more specialized glyptosternoid genera, such as Pareuchiloglanis, originated in the Pleistocene (within 1.9 Mya). The specia- tion of glyptosternoid fishes is hypothesized to be closely related with the uplift of the Qinghai-Tibet Plateau.