Lycodon and Dinodon: One Genus Or Two? Evidence from Molecular Phylogenetics and Morphological Comparisons ⇑ Peng Guo A,B, , Liang Zhang C, Qin Liu A,B, Cao Li B,D, R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

AMANI NATURE RESERVE an Introduction
Field Guides AMANI NATURE RESERVE an introduction This guide was developed to help participants on Tropical Biology Association field courses to learn about the Amani Nature Reserve and the forests of the East Usambara Mountains. It includes an introduction to the East Usambaras and describes the ecology, flora and fauna of the area. The history of management and conservation of the Amani Nature Reserve, together with its current status, is outlined. This publication was funded by the European Commission (B7-6200/01/0370/ENV). For any queries concerning this document please contact: Tropical Biology Association Department of Zoology Downing Street, Cambridge CB2 3EJ United Kingdom Tel: +44 (0) 1223 336619 e-mail: [email protected] © Tropical Biology Association 2007 A Banson production Printed by Swaingrove Field Guides AMANI NATURE RESERVE an introduction TBA Field Guide CONTENTS EAST USAMBARA MOUNTAINS 3 Geographical history 3 Flora and fauna of the Usambara Mountains 3 Human impacts 3 History of Amani 5 History of Amani Botanical Garden 5 FLORA OF THE EASTERN USAMBARAS & AMANI 6 Vegetation cover of the East Usambara Mountains 6 Endemic plants in Amani 7 Introduced (alien and invasive) species 7 Case study of an introduced species: Maesopsis eminii (Rhamnaceae) 8 FAUNA OF AMANI 9 Vertebrates 9 Invertebrates 13 MANAGEMENT OF AMANI NATURE RESERVE 14 Conservation 14 REFERENCES 16 2 Amani Nature Reserve EAST USAMBARA MOUNTAINS An overview Geographical history The Amani Nature Reserve is located in the East Usambara region. This is part of the Eastern Arc Mountains, an isolated mountain chain of ancient crystalline rock formed through a cycle of block faulting and erosion that stretches from the Taita Hills in Kenya down to the Southern Highlands in Tanzania. -

New Records of Snakes (Squamata: Serpentes) from Hoa Binh Province, Northwestern Vietnam
Bonn zoological Bulletin 67 (1): 15–24 May 2018 New records of snakes (Squamata: Serpentes) from Hoa Binh Province, northwestern Vietnam Truong Quang Nguyen1,2,*, Tan Van Nguyen 1,3, Cuong The Pham1,2, An Vinh Ong4 & Thomas Ziegler5 1 Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 2 Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet Road, Hanoi, Vietnam 3 Save Vietnam’s Wildlife, Cuc Phuong National Park, Ninh Binh Province, Vietnam 4 Vinh University, 182 Le Duan Road, Vinh City, Nghe An Province, Vietnam 5 AG Zoologischer Garten Köln, Riehler Strasse 173, D-50735 Cologne, Germany * Corresponding author. E-mail: [email protected] Abstract. We report nine new records of snakes from Hoa Binh Province based on a reptile collection from Thuong Tien, Hang Kia-Pa Co, Ngoc Son-Ngo Luong nature reserves, and Tan Lac District, comprising six species of Colubri- dae (Dryocalamus davisonii, Euprepiophis mandarinus, Lycodon futsingensis, L. meridionalis, Sibynophis collaris and Sinonatrix aequifasciata), one species of Pareatidae (Pareas hamptoni) and two species of Viperidae (Protobothrops mu- crosquamatus and Trimeresurus gumprechti). In addition, we provide an updated list of 43 snake species from Hoa Binh Province. The snake fauna of Hoa Binh contains some species of conservation concern with seven species listed in the Governmental Decree No. 32/2006/ND-CP (2006), nine species listed in the Vietnam Red Data Book (2007), and three species listed in the IUCN Red List (2018). Key words. New records, snakes, taxonomy, Hoa Binh Province. -

Occurrence and Distribution of Snake Species in Balochistan Province, Pakistan
Pakistan J. Zool., pp 1-4, 2021. DOI: https://dx.doi.org/10.17582/journal.pjz/20181111091150 Short Communication Occurrence and Distribution of Snake Species in Balochistan Province, Pakistan Saeed Ahmed Essote1, Asim Iqbal1, Muhammad Kamran Taj2*, Asmatullah Kakar1, Imran Taj2, Shahab-ud-Din Kakar1 and Imran Ali 3,4* 1Department of Zoology, University of Balochistan, Quetta 2Center for Advanced Studies in Vaccinology and Biotechnology, University of Balochistan, Quetta, Pakistan. 3Institute of Biochemistry, University of Balochistan, Quetta 4 Plant Biomass Utilization Research Unit, Chulalongkorn University, Bangkok, 10330, Article Information Thailand. Received 11 November 2018 Revised 11 October 2020 Accepted 10 December 2020 ABSTRACT Available online 28 April 2021 Authors’ Contribution The current study was conducted in Zhob, Quetta, Sibi, Kalat, Naseer Abad and Makran Divisions of SAE carried the research with the Balochistan Province. A total of 619 snake specimens representing 6 families, 20 genera and 37 species help of other authors and wrote were collected. The family wise representation among collected specimens has been Boidae (4.6%), the manuscript. AI, MKT and IT Leptotyphlopidae (7.5%), Typhlopidae (10.3%), Elapidae (11.7%), Viperidae (13.4%) and Colubridae helped in the experimental work. AK (52.5%). The percentage of family Boidae, Typhlopidae, Elapidae and Leptotyphlopidae were high in and SDK classified the species and Sibi Division while family Viperidae and Colubridae were dominant in Quetta Division. The family proofread the article. IA helped in arranging contents of the article. Colubridae has been the most dominant in the Province, having ten genera viz., Boiga (6.8%) Coluber (10.1%), Eirenis (2.5%), Lycodon (3.5%), Lytorhynchus (6.1%), Oligodon (4.7%), Natrix (1.7%), (7.5%), Key words Ptyas (2.9 %) Spalerosophis (6.3%) and Psammophis. -

Cfreptiles & Amphibians
HTTPS://JOURNALS.KU.EDU/REPTILESANDAMPHIBIANSTABLE OF CONTENTS IRCF REPTILES & AMPHIBIANSREPTILES • VOL &15, AMPHIBIANS NO 4 • DEC 2008 • 28(1):157–158189 • APR 2021 IRCF REPTILES & AMPHIBIANS CONSERVATION AND NATURAL HISTORY TABLE OF CONTENTS FEATUREPredation ARTICLES on a Common Wolfsnake, . Chasing Bullsnakes (Pituophis catenifer sayi) in Wisconsin: LycodonOn the Road to aulicusUnderstanding the Ecology (Colubridae),and Conservation of the Midwest’s Giant Serpent ...................... by anJoshua M. KapferIndian 190 . The Shared History of Treeboas (Corallus grenadensis) and Humans on Grenada: Roller,A Hypothetical Coracias Excursion ............................................................................................................................ benghalensis (Coraciidae),Robert W. Henderson 198 RESEARCH ARTICLES in. The the Texas Horned Sathyamangalam Lizard in Central and Western Texas ....................... Emily Henry, JasonTiger Brewer, Krista Mougey, Reserve, and Gad Perry 204 . The Knight Anole (Anolis equestris) in Florida .............................................TamilBrian J. Camposano, Kenneth Nadu, L. Krysko, Kevin M. Enge,India Ellen M. Donlan, and Michael Granatosky 212 CONSERVATION ALERT . World’s Mammals in Crisis ...............................................................................................................................Sreedharan Nair Vishnu and Chinnasamy Ramesh .............................. 220 . More Than Mammals ..................................................................................................................................................................... -

Genus Lycodon)
Zoologica Scripta Multilocus phylogeny reveals unexpected diversification patterns in Asian wolf snakes (genus Lycodon) CAMERON D. SILER,CARL H. OLIVEROS,ANSSI SANTANEN &RAFE M. BROWN Submitted: 6 September 2012 Siler, C. D., Oliveros, C. H., Santanen, A., Brown, R. M. (2013). Multilocus phylogeny Accepted: 8 December 2012 reveals unexpected diversification patterns in Asian wolf snakes (genus Lycodon). —Zoologica doi:10.1111/zsc.12007 Scripta, 42, 262–277. The diverse group of Asian wolf snakes of the genus Lycodon represents one of many poorly understood radiations of advanced snakes in the superfamily Colubroidea. Outside of three species having previously been represented in higher-level phylogenetic analyses, nothing is known of the relationships among species in this unique, moderately diverse, group. The genus occurs widely from central to Southeast Asia, and contains both widespread species to forms that are endemic to small islands. One-third of the diversity is found in the Philippine archipelago. Both morphological similarity and highly variable diagnostic characters have contributed to confusion over species-level diversity. Additionally, the placement of the genus among genera in the subfamily Colubrinae remains uncertain, although previous studies have supported a close relationship with the genus Dinodon. In this study, we provide the first estimate of phylogenetic relationships within the genus Lycodon using a new multi- locus data set. We provide statistical tests of monophyly based on biogeographic, morpho- logical and taxonomic hypotheses. With few exceptions, we are able to reject many of these hypotheses, indicating a need for taxonomic revisions and a reconsideration of the group's biogeography. Mapping of color patterns on our preferred phylogenetic tree suggests that banded and blotched types have evolved on multiple occasions in the history of the genus, whereas the solid-color (and possibly speckled) morphotype color patterns evolved only once. -

Annual Review 2015
ANNUAL REVIEW 2015 01 Our mission 02 President’s statement 03 Chairman’s statement 05 Chief Executive’s statement 07 Conservation 08 Conservation science 10 Out in the field 12 Community conservation 14 Engagement 16 RZSS Edinburgh Zoo 20 2015 highlights 22 RZSS Highland Wildlife Park 26 Get involved 28 Financial summary 30 Our people and structure 31 Board, fellows and patrons 33 RZSS Edinburgh Zoo Inventory 38 RZSS Highland Wildlife Park Inventory 40 About us Front cover: Arktos the polar bear at the Highland Wildlife Park, taken by RZSS Photographer in Residence Laurie Campbell OUR MISSION Connecting people with nature. Safeguarding species from extinction. The Budongo Conservation Field Station in Uganda celebrated its 25th anniversary in 2015 Annual Review 2015 01 1 Victoria, the UK’s only female polar PRESIDENT’S bear, who arrived at the Highland Wildlife Park in March STATEMENT 2 Jayendra and Roberta, our pair of endangered Asiatic lions, were introduced to one another in April After nearly ten years, it is strange to be writing my final foreword for the Royal Zoological Society of Scotland’s Annual Review. On my first day, I spoke of Secondly, there is the respect Looking to the future, I encourage the privilege I felt to be your for and care of our animals. trustees, staff and members to President and that sentiment I have always recognised that retain their passion for our vision still remains. Therefore, there this is an essential part of our and to be ready, on occasion, to is a lump in my throat as I pen DNA and the exemplary record take measured risks. -

Herp. Bulletin 101.Qxd
Mountain wolf snake ( Lycodon r . ruhstrati ) predation on an exotic lizard, Anolis sagrei , in Chiayi County, Taiwan GERRUT NORVAL 1, SHAO-CHANG HUANG 2 and JEAN-JAY MAO 3 1 Applied Behavioural Ecology & Ecosystem Research Unit, Department of Nature Conservation, UNISA, Private Bag X6, Florida, 1710, Republic of South Africa . Email: [email protected] [author for correspondence] 2 Department of Life Science, Tunghai University. No. 181, Sec. 3, Taichung-Kan Road, Taichung, 407- 04, Taiwan, R.O.C . 3 Department of Natural Resources, National I-Lan University, No. 1, Shen-Lung Rd., Sec. 1, I-Lan City 260, Taiwan, R.O.C . ABSTRACT – The Mountain wolf snake ( Lycodon ruhstrati ruhstrati ) is a common snake species at low elevations all over Taiwan. Still, it appears to be poorly studied in Taiwan and adjacent areas since little has been reported about this species. On 26 th August 2002 ten L. r. ruhstrati eggs were obtained from an adult female, one of two that were caught a day before, and eight of the eggs hatched successfully on 14 th October 2002. While in captivity all the adults preyed upon Anolis sagrei , which were given to them as prey, while two neonates accepted A. sagrei hatchlings offered to them as food. On February 18 th , 2006, a DOR Mountain wolf snake, with an A. sagrei in its stomach, was found on a tarred road in Santzepu, Sheishan District, Chiayi County. This appears to be the first report from Taiwan of the Mountain wolf snake ( L. r. ruhstrati ) preying on the exotic introduced lizard A. -

TFCG Technical Paper 18 the VERTEBRATE BIODIVERSITY AND
TFCG Technical Paper 18 THE VERTEBRATE BIODIVERSITY AND FOREST CONDITION OF UDZUNGWA MOUNTAIN FORESTS IN MUFINDI DISTRICT By N. Doggart, C. Leonard, A. Perkin, M. Menegon and F. Rovero Dar es Salaam June 2008 Cover photographs by Michele Menegon. From left to right. 1. Horned bush viper eating a reed frog. 2. View of Igoda Forest and adjacent tea fields. 3. Spiny flanked chameleon (Chamaeleo laterispinis) ¤ Tanzania Forest Conservation Group Suggested citations: Whole report Doggart, N., C. Leonard, A. Perkin, M. Menegon and F. Rovero (2008). The Biodiversity and forest condition of Udzungwa Mountain forests in Mufindi District. TFCG Technical Paper No 18. DSM, Tz. 1- 142 pp. Sections with Report: (example using section 3) Menegon, M., (2008). Reptiles and Amphibians. In: Doggart, N., C. Leonard, A. Perkin, M. Menegon and F. Rovero (2008). The Biodiversity and forest condition of Udzungwa Mountain forests in Mufindi District.TFCG Technical Paper No 18. DSM, Tz. 1 - 142 pp. EXECUTIVE SUMMARY Introduction The Eastern Arc Mountain forests in Mufindi District lie at the south-western extreme of the Eastern Arc. The forests are found on the Mufindi plateau at the top of the Mufindi escarpment. The plateau includes a mosaic of forest, commercial tea cultivation, pine and eucalyptus plantation, coffee and subsistence agriculture. The forests of Mufindi are highly fragmented and many of them show high levels of disturbance some of which dates back over 50 years. Lovett & Pócs (1993) suggest that parts of the larger Mufindi forests such as Kigogo were cultivated in the mid 19th Century and cite the presence of agricultural ridges under the forest. -

New Country Records of Reptiles from Laos
Biodiversity Data Journal 1: e1015 doi: 10.3897/BDJ.1.e1015 Taxonomic paper New country records of reptiles from Laos Vinh Quang Luu†,‡, Truong Quang Nguyen§,|, Thomas Calame¶, Tuoi Thi Hoang#, Sisomphone Southichack††, Michael Bonkowski|, Thomas Ziegler‡,| † Department of Wildlife, Faculty of Natural Resource and Environmental Management, Vietnam Forestry University, Xuan Mai, Chuong My, Hanoi, Vietnam ‡ AG Zoologischer Garten Köln, Riehler Strasse 173, D-50735 Cologne, Germany § Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Hanoi, Vietnam | Zoological Institute, University of Cologne, Zülpicher Strasse 47b, D-50674 Cologne, Germany ¶ WWF Greater Mekong, House No. 39, Unit 05, Ban Saylom, Vientiane, Lao People's Democratic Republic # Biodiversity Center, Faculty of Natural Resource and Environmental Management, Vietnam Forestry University, Xuan Mai, Chuong My, Hanoi, Vietnam †† Hin Nam No National Protected Area, Boualapha District, Khammouane Province, Lao People's Democratic Republic Corresponding author: Vinh Quang Luu ([email protected]) Academic editor: Johannes Penner Received: 27 Oct 2013 | Accepted: 04 Dec 2013 | Published: 10 Dec 2013 Citation: Luu V, Nguyen T, Calame T, Hoang T, Southichack S, Bonkowski M, Ziegler T (2013) New country records of reptiles from Laos. Biodiversity Data Journal 1: e1015. doi: 10.3897/BDJ.1.e1015 Abstract Four species of reptiles, of which one is represented by one of its subspecies, are recorded for the first time from Laos: Cyrtodactylus phongnhakebangensis, Lycodon futsingensis, and L. ruhstrati, as L. ruhstrati abditus, from limestone forests in Khammouane Province and Cyrtodactylus pseudoquadrivirgatus from hill evergreen forest in Salavan Province. These discoveries of lizards and snakes bring the total species number of reptiles to 189 in Laos. -

The Amphibians and Reptiles of Kakamega Forest, Western Kenya
07_Wagner&Böhme-2.qxd 12.08.2007 14:01 Uhr Seite 123 Bonner zoologische Beiträge Band 55 (2006) Heft 2 Seiten 123–150 Bonn, Juli 2007 Herpetofauna Kakamegensis – The amphibians and reptiles of Kakamega Forest, western Kenya Philipp WAGNER & Wolfgang BÖHME Bonn, Germany Abstract. We present an annotated checklist of the herpetofauna of Kakamega Forest with comments on the biology and systematics of the taxa. Twenty-five amphibian, one turtle, 22 lizard and 36 snake species are recorded from within the forest and its immediate environment. We discuss the generalized zoogeography of the forest and distribution pattern of the taxa comment on the protection of the forest. Analysis of the reptile species composition shows Kakamega Forest to be similar to the Guinea-Congolian rainforest and is considered the easternmost remnant of this forest block. Kakamega forest has a high diversity value for Kenya and represents a diversity hotspot on a national scale. Two species, Lygodac- tylus gutturalis and Psammophis phillipsi, are recorded in Kenya for the first time. Several other first records and the description of a new species (Agamidae: Agama finchi) were published already separately. Keywords. East Africa, Kenya, Kakamega Forest, herpetological survey, checklist, national diversity hotspot. 1. INTRODUCTION In Africa tropical rainforests extend from southern Sene- covery and scientific descriptions. Uganda, for example gal in the west to the coastal forests of Kenya and Tanza- has lost 86 % of its original forest in the past two decades nia in the east (COLLINS 1992). The East African rainforests and the remaining parts are isolated fragments (VONESH belong to different biogeographical clades. -
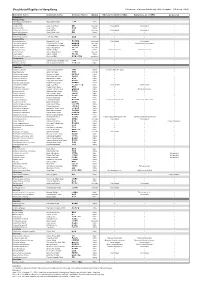
Checklist of Reptiles in Hong Kong © Programme of Ecology & Biodiversity, HKU (Last Update: 10 September 2012)
Checklist of Reptiles in Hong Kong © Programme of Ecology & Biodiversity, HKU (Last Update: 10 September 2012) Scientific name Common name Chinese Name Status Ades & Kendrick (2004) Karsen et al. (1998) Uetz et al. Testudines Platysternidae Platysternon megacephalum Big-headed Terrapin 大頭龜 Native Cheloniidae Caretta caretta Loggerhead Turtle 蠵龜 Uncertain Not included Not included Chelonia mydas Green Turtle 緣海龜 Native Eretmochelys imbricata Hawksbill Turtle 玳瑁 Uncertain Not included Not included Lepidochelys olivacea Pacific Ridley Turtle 麗龜 Native Dermochelyidae Dermochelys coriacea Leatherback Turtle 稜皮龜 Native Geoemydidae Cuora amboinensis Malayan Box Turtle 馬來閉殼龜 Introduced Not included Not included Cuora flavomarginata Yellow-lined Box Terrapin 黃緣閉殼龜 Uncertain Cistoclemmys flavomarginata Cuora trifasciata Three-banded Box Terrapin 三線閉殼龜 Native Mauremys mutica Chinese Pond Turtle 黃喉水龜 Uncertain Mauremys reevesii Reeves' Terrapin 烏龜 Native Chinemys reevesii Chinemys reevesii Ocadia sinensis Chinese Striped Turtle 中華花龜 Uncertain Sacalia bealei Beale's Terrapin 眼斑水龜 Native Trachemys scripta elegans Red-eared Slider 巴西龜 / 紅耳龜 Introduced Trionychidae Palea steindachneri Wattle-necked Soft-shelled Turtle 山瑞鱉 Uncertain Pelodiscus sinensis Chinese Soft-shelled Turtle 中華鱉 / 水魚 Native Squamata - Serpentes Colubridae Achalinus rufescens Rufous Burrowing Snake 棕脊蛇 Native Achalinus refescens (typo) Ahaetulla prasina Jade Vine Snake 綠瘦蛇 Uncertain Amphiesma atemporale Mountain Keelback 無顳鱗游蛇 Native -

P. 1 AC27 Inf. 7 (English Only / Únicamente En Inglés / Seulement
AC27 Inf. 7 (English only / únicamente en inglés / seulement en anglais) CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________ Twenty-seventh meeting of the Animals Committee Veracruz (Mexico), 28 April – 3 May 2014 Species trade and conservation IUCN RED LIST ASSESSMENTS OF ASIAN SNAKE SPECIES [DECISION 16.104] 1. The attached information document has been submitted by IUCN (International Union for Conservation of * Nature) . It related to agenda item 19. * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. The responsibility for the contents of the document rests exclusively with its author. AC27 Inf. 7 – p. 1 Global Species Programme Tel. +44 (0) 1223 277 966 219c Huntingdon Road Fax +44 (0) 1223 277 845 Cambridge CB3 ODL www.iucn.org United Kingdom IUCN Red List assessments of Asian snake species [Decision 16.104] 1. Introduction 2 2. Summary of published IUCN Red List assessments 3 a. Threats 3 b. Use and Trade 5 c. Overlap between international trade and intentional use being a threat 7 3. Further details on species for which international trade is a potential concern 8 a. Species accounts of threatened and Near Threatened species 8 i. Euprepiophis perlacea – Sichuan Rat Snake 9 ii. Orthriophis moellendorfi – Moellendorff's Trinket Snake 9 iii. Bungarus slowinskii – Red River Krait 10 iv. Laticauda semifasciata – Chinese Sea Snake 10 v.