In Situ Proximity Ligation Assay Protocols
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analytically Sensitive Protein Detection in Microtiter Plates By
Papers in Press. Published July 18, 2017 as doi:10.1373/clinchem.2017.271833 The latest version is at http://hwmaint.clinchem.aaccjnls.org/cgi/doi/10.1373/clinchem.2017.271833 Clinical Chemistry 63:9 Molecular Diagnostics and Genetics 000–000 (2017) Analytically Sensitive Protein Detection in Microtiter Plates by Proximity Ligation with Rolling Circle Amplification Tonge Ebai,1 Felipe Marques Souza de Oliveira,1 Liza Löf,1 Lotta Wik,1 Caroline Schweiger,2,4 Anders Larsson,3 Ulrich Keilholtz,4 Johannes Haybaeck,2,5 Ulf Landegren,1*† and Masood Kamali-Moghaddam1*† BACKGROUND: Detecting proteins at low concentrations tine instrumentation available in hospitals and research in plasma is crucial for early diagnosis. Current tech- laboratories. niques in clinical routine, such as sandwich ELISA, pro- © 2017 American Association for Clinical Chemistry vide sensitive protein detection because of a dependence on target recognition by pairs of antibodies, but detection of still lower protein concentrations is often called for. Antibody-based protein detection assays are valuable tools Proximity ligation assay with rolling circle amplification for clinical diagnostics and research applications. However, (PLARCA) is a modified proximity ligation assay (PLA) for analytically specific and sensitive protein detection via the molecular complexity and wide protein concentration binding of target proteins by 3 antibodies, and signal ranges of biofluids such as plasma (1) complicate detection. amplification via rolling circle amplification (RCA) in The challenge of specific detection increases rapidly with microtiter wells, easily adapted to instrumentation in use decreasing concentrations of proteins of interest. In partic- in hospitals. ular, early detection of disease via leakage markers specifi- cally emanating from affected tissues places stringent de- mands on both the analytical specificity and sensitivity. -

Streamlined Circular Proximity Ligation Assay Provides High Stringency and Compatibility with Low-Affinity Antibodies
Streamlined circular proximity ligation assay provides PNAS PLUS high stringency and compatibility with low-affinity antibodies Roxana Jalilia,b, Joe Horeckaa,1, James R. Swartzb, Ronald W. Davisa,2, and Henrik H. J. Perssona,2 aStanford Genome Technology Center, Stanford University School of Medicine, Palo Alto, CA 94304; and bDepartment of Chemical Engineering, Stanford University, Stanford, CA 94305 Contributed by Ronald W. Davis, December 7, 2017 (sent for review October 20, 2017; reviewed by Simon Fredriksson and S. Patrick Walton) Proximity ligation assay (PLA) is a powerful tool for quantitative probes are individually conjugated to short ssDNA molecules to detection of protein biomarkers in biological fluids and tissues. form proximity probes that carry either a phosphorylated 5′ end Here, we present the circular proximity ligation assay (c-PLA), a or a 3′-hydroxyl group. When the probe pairs subsequently bind highly specific protein detection method that outperforms tradi- to their cognate target analyte in solution, the associated DNA tional PLA in stringency, ease of use, and compatibility with low- strands are brought into close proximity and aligned by hybrid- affinity reagents. In c-PLA, two proximity probes bind to an ization to a third bridging oligonucleotide. The free DNA ends analyte, providing a scaffolding that positions two free oligonu- are ligated, forming a new DNA sequence that is amplified and cleotides such that they can be ligated into a circular DNA quantified using qPCR (Fig. 1A). We have previously showed molecule. This assay format stabilizes antigen proximity probe that PLA provides femtomolar sensitivity and a wide dynamic complexes and enhances stringency by reducing the probability of range over five orders of magnitude, consuming as little as 1 μL random background ligation events. -

Detecting Individual Extracellular Vesicles Using a Multicolor in Situ
www.nature.com/scientificreports OPEN Detecting individual extracellular vesicles using a multicolor in situ proximity ligation assay with flow Received: 28 April 2016 Accepted: 09 September 2016 cytometric readout Published: 29 September 2016 Liza Löf 1, Tonge Ebai1, Louise Dubois2, Lotta Wik1, K. Göran Ronquist2, Olivia Nolander1, Emma Lundin1, Ola Söderberg1, Ulf Landegren1 & Masood Kamali-Moghaddam1 Flow cytometry is a powerful method for quantitative and qualitative analysis of individual cells. However, flow cytometric analysis of extracellular vesicles (EVs), and the proteins present on their surfaces has been hampered by the small size of the EVs – in particular for the smallest EVs, which can be as little as 40 nm in diameter, the limited number of antigens present, and their low refractive index. We addressed these limitations for detection and characterization of EV by flow cytometry through the use of multiplex and multicolor in situ proximity ligation assays (in situ PLA), allowing each detected EV to be easily recorded over background noise using a conventional flow cytometer. By targeting sets of proteins on the surface that are specific for distinct classes of EVs, the method allows for selective recognition of populations of EVs in samples containing more than one type of EVs. The method presented herein opens up for analyses of EVs using flow cytometry for their characterization and quantification. Cells have the capacity to release different kinds of vesicles into the extracellular space, collectively named extra- cellular vesicles (EVs). EVs can be grossly subdivided into three subclasses based on their biogenesis and in order of increasing sizes; exosomes, microvesicles, and apoptotic bodies. -

Proximity Ligation Assay Detection of Protein–DNA Interactions—Is There a Link Between Heme Oxygenase-1 and G-Quadruplexes?
antioxidants Article Proximity Ligation Assay Detection of Protein–DNA Interactions—Is There a Link between Heme Oxygenase-1 and G-quadruplexes? Wojciech Krzeptowski 1,†, Patryk Chudy 1,†, Grzegorz Sokołowski 1, Monika Zukowska˙ 1, Anna Kusienicka 1 , Agnieszka Seretny 1, Agata Kalita 2 , Alicja Czmoczek 1, Jakub Gubała 1 , Sonia Baran 1, Damian Klóska 1, Mateusz Jez˙ 1, Jacek St˛epniewski 1, Krzysztof Szade 1, Agata Szade 1 , Anna Grochot-Prz˛eczek 1 , Alicja Józkowicz 1 and Witold N. Nowak 1,* 1 Department of Medical Biotechnology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, 30387 Kraków, Poland; [email protected] (W.K.); [email protected] (P.C.); [email protected] (G.S.); [email protected] (M.Z.);˙ [email protected] (A.K.); [email protected] (A.S.); [email protected] (A.C.); [email protected] (J.G.); [email protected] or [email protected] (S.B.); [email protected] (D.K.); [email protected] (M.J.); [email protected] (J.S.); [email protected] (K.S.); [email protected] (A.S.); [email protected] (A.G.-P.); [email protected] (A.J.) 2 Department of General Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, 30387 Kraków, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-126646024 † Equally contributed as first authors. Abstract: G-quadruplexes (G4) are stacked nucleic acid structures that are stabilized by heme. -

Proximity Ligation
! " # $% &'( )*)&+'( ,-, (&.*)&-,-& ,"&*(& ,", // Dissertation for the Degree of Doctor of Philosophy (Faculty of Medicine) in Molecular Medicine presented at Uppsala University in 2002 ABSTRACT Fredriksson, S. 2002. Proximity Ligation. Transforming protein analysis into nucleic acid detection through proximity-dependent ligation of DNA sequence tagged protein-binders. Acta Universitatis Upsaliensis. Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1192. 34 pp. Uppsala. ISBN 91-554-5426-7 A novel technology for protein detection, proximity ligation, has been developed along with improved methods for in situ synthesis of DNA microarrays. Proximity ligation enables a specific and quantitative transformation of proteins present in a sample into nucleic acid sequences. As pairs of so-called proximity probes bind the individual target protein molecules at distinct sites, these reagents are brought in close proximity. The probes consist of a protein specific binding part coupled to an oligonucleotide with either a free 3’- or 5’-end capable of hybridizing to a common connector oligonucleotide. When the probes are in proximity, promoted by target binding, then the DNA strands can be joined by enzymatic ligation. The nucleic acid sequence that is formed can then be amplified and quantitatively detected in a real-time monitored polymerase chain reaction. This convenient assay is simple to perform and allows highly sensitive protein detection. Parallel analysis of multiple proteins by DNA microarray technology is anticipated for proximity ligation and enabled by the information carrying ability of nucleic acids to define the individual proteins. Assays detecting cytokines using SELEX aptamers or antibodies, monoclonal and polyclonal, are presented in the thesis. Microarrays synthesized in situ using photolithographic methods generate impure products due to damaged molecules and interrupted synthesis. -

Optimization of Proximity Ligation Assay Based Western Blotting
UPTEC X11 036 Examensarbete 30 hp November 2011 Optimization of proximity ligation assay based Western blotting Johan Johansson Molecular Biotechnology Programme Uppsala University School of Engineering UPTEC X 11 036 Date of issue 2011-11 Author Johan Johansson Title (English) Optimization of proximity ligation assay based Western blotting Title (Swedish) Abstract Many of today’s methods for the detection of biomolecules suffer from a high limit of detection due to poor signal generation upon recognition of target. By applying and optimizing proximity ligation assay (PLA) in Western blotting (WB), the limit of detection has been lowered down to the picomolar range. In this report I have optimized the different parameters that affect the signal generation and explored possibilities to increase the ease of use, by merging protocol steps and performing signal generating reactions at room temperature. Keywords Western blotting, proximity ligation assay, signal amplification, specificity, optimization Supervisors Åsa Hagner-McWhirter, GE Healthcare Scientific reviewer Anna Edman-Örlefors, GE Healthcare Project name Sponsors Language Security English Classification ISSN 1401-2138 Supplementary bibliographical information Pages 30 Biology Education Centre Biomedical Center Husargatan 3 Uppsala Box 592 S-75124 Uppsala Tel +46 (0)18 4710000 Fax +46 (0)18 471 4687 Optimization of proximity ligation assay based Western blotting Johan Johansson Populärvetenskaplig sammanfattning I dag kan många sjukdomar botas redan innan de brutit ut. Genom att dra nytta av så kallade biomarkörer, molekyler som kan visa på en specifik sjukdom så kan man starta en behandling före symptom uppstår. Det är dock svårt att diagnostisera med hjälp av biomarkörer. Detta då det finns väldigt lite av dem, samt att det är svårt att specifikt detektera en viss sort av dem. -

Hipla: High-Throughput Imaging Proximity Ligation Assay
bioRxiv preprint doi: https://doi.org/10.1101/371062; this version posted July 17, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. HiPLA: High-throughput imaging Proximity Ligation Assay Leonid A. Serebryannyy1 and Tom Misteli1 1 Cell Biology of Genomes Group, National Cancer Institute, NIH, Building 41, 41 Library Drive, Bethesda, MD 20892, USA Correspondence to [email protected] Keywords: proximity ligation, high-throughput imaging, Hutchinson-Gilford progeria syndrome, lamin proteomics bioRxiv preprint doi: https://doi.org/10.1101/371062; this version posted July 17, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: Protein-protein interactions are essential for cellular structure and function. To delineate how the intricate assembly of protein interactions contribute to cellular processes in health and disease, new methodologies that are both highly sensitive and can be applied at large scale are needed. Here, we develop HiPLA (high-throughput imaging proximity ligation assay), a method that employs the antibody-based proximity ligation assay in a high-throughput imaging screening format to systematically probe protein interactomes. Using HiPLA, we probe the interaction of 60 proteins and associated PTMs with the nuclear lamina in a model of the premature aging disorder Hutchinson- Gilford progeria syndrome (HGPS). We identify a subset of proteins that differentially interact with the nuclear lamina in HGPS. In combination with quantitative indirect immunofluorescence, we find that the majority of differential interactions were accompanied by corresponding changes in expression of the interacting protein. -

Technology Development in the Field of Ligand Binding Assays Comparison Between ELISA and Other Methods
20-X5 Technology Development inthe Field of Ligand Binding Assays Comparison between ELISA and other methods Tanya Al-Khafaf, Björn Ancker Persson, Johanna Cederblad, Albert Häggström, Reneh Kostines, Lina Löfström, Ella Schleimann-Jensen Client: Mercodia AB Client representative: Henning Henschel Supervisor: Lena Henriksson 1MB332, Independent Project in Molecular Biotechnology, 15 hp, spring semester 2020 Master Programme in Molecular Biotechnology Engineering Biology Education Centre, Uppsala University Abstract In this project, given to us by Mercodia AB, research in the field of im- munoassays is done in order to investigate if there are methods that are better than the conventional ELISA. ELISA is known to have some issues, such as ”The Hook effect”, many washing steps and cross-reactivity with the antibod- ies used in the assay. Therefore the need of other methods has arised. The result of the research showed that there are a huge number of methods that measure specific biomarkers. In this report 17 different techniques are presented. These techniques are: Mass Spectrometry (MS), Chemilumines- cence Immunoassay (CLIA), AlphaLISA, Lateral Flow Immunoassay (LFIA), Microfluidics-based Immunoassays, Paper Based Immunoassays, Biosensors and Aptasensors, Immuno-PCR, Proximity Ligand Assay (PLA), Proximity Extension Assays (PEA), Meso-scale discovery (MSD), Multiplex Assay, Dig- ital Bioassay, Bioluminescence Resonance Emission Transfer (BRET), Homo- geneous Time Resolved Fluorescence (HTRF) and NanoBiT. Each of the listed methods are compared according to several parameters such as specificity, sen- sitivity, measure range, sample volume, degree of automation, runtime and cost for each analyzed sample. The methods that showed an upward trend were: AlphaLISA, BRET, Biosen- sors, CLIA, Digital ELISA, methods using gold nanoparticles (AuNPs), HTRF, Immuno-PCR, Lateral Flow, MSD, Microfluidics, Multiplex methods, NanoBiT, paper-based, PEA, Simoa and Single molecule detection. -

Padlock and Proximity Probes for in Situ and Array-Based Analyses: Tools for the Post-Genomic Era
Comparative and Functional Genomics Comp Funct Genom 2003; 4: 525–530. Published online in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/cfg.326 Conference Review Padlock and proximity probes for in situ and array-based analyses: tools for the post-genomic era Ulf Landegren*, Fredrik Dahl, Mats Nilsson, Simon Fredriksson, Johan Baner,´ Mats Gullberg, Jonas Jarvius, Sigrun Gustafsdottir, Ola Soderberg,¨ Olle Ericsson, Johan Stenberg and Edith Schallmeiner Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, Sweden *Correspondence to: Abstract Ulf Landegren, Department of Genetics and Pathology, Rudbeck Highly specific high-throughput assays will be required to take full advantage of Laboratory, Uppsala University, the accumulating information about the macromolecular composition of cells and Sweden. tissues, in order to characterize biological systems in health and disease. We discuss E-mail: the general problem of detection specificity and present the approach our group [email protected] has taken, involving the reformatting of analogue biological information to digital reporter segments of genetic information via a series of DNA ligation assays. The assays enable extensive, coordinated analyses of the numbers and locations of genes, Received: 6 June 2003 transcripts and protein. Copyright 2003 John Wiley & Sons, Ltd. Revised: 5 August 2003 Accepted: 5 August 2003 A background to molecular analyses core, the problem of analysing macromolecules in biological samples is one of specificity of detection, Complete genome sequences are becoming avail- and the requirements are extreme: the two double- able for an increasing number of organisms, allow- stranded haploid genomes in any human interphase ing research on the corresponding species to tran- cell together comprise approximately 13 billion sit to a post-genomic phase. -
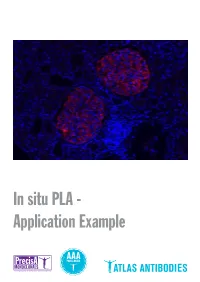
In Situ PLA - Application Example Detection of Protein-Protein Interactions by in Situ PLA with Precisa Monoclonals and Triple a Polyclonals
In situ PLA - Application Example Detection of protein-protein interactions by in situ PLA with PrecisA Monoclonals and Triple A Polyclonals Proteins are complex biological mole- protein-protein interactions, as well as for recombinant antigen as affinity ligand for cules essential for cellular structure and detection of individual proteins and pro- the Triple A Polyclonals. PrecisA Mono- functions. The majority of proteins com- tein modifications in cells or tissue sec- clonals are purified on protein A columns. monly interact with various molecules, tions. Importantly, the antibody specificity is vali- including other proteins, in order to exert For achieving reliable results with the PLA dated by an extensive IHC analysis in 44 their functions. These protein-protein in- assay, the choice of primary antibodies normal and 20 cancerous human tissues. teractions are involved in a wide range of is crucial. Only specific and selective an- As mentioned above, the main advan- cellular processes, including e.g. protein tibodies will enable a successful PLA re- tages of is PLA in comparison to other modifications, transport, signaling and action. General requirements for the pri- techniques is the possibility to detect pro- cell cycling. mary antibodies are that they should be tein-protein interactions with subcellular Studies of protein interactions are criti- of IgG class, affinity-purified and not least, resolution in native tissue sections or cells cal for the characterization of protein target-specific. In addition, for the detec- using microscopy. This can be particularly functions in the context of cell biology in tion of protein-protein interaction, the two important in e.g. -
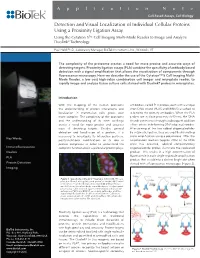
Detection and Visual Localization of Individual Cellular Proteins Using a Proximity Ligation Assay
Application Note Cell-Based Assays, Cell Biology Detection and Visual Localization of Individual Cellular Proteins Using a Proximity Ligation Assay Using the Cytation 5™ Cell Imaging Multi-Mode Reader to Image and Analyze ® Duolink Technology Paul Held Ph.D., Laboratory Manager, BioTek Instruments, Inc., Winooski, VT The complexity of the proteome creates a need for more precise and accurate ways of detecting targets. Proximity ligation assays (PLA) combine the specificity of antibody based detection with a signal amplification that allows the visualization of components through fluorescence microscopy. Here we describe the use of the Cytation™ 5 Cell Imaging Multi- Mode Reader, a low cost high-value combination cell imager and microplate reader, to rapidly image and analyze tissue culture cells stained with Duolink® probes in microplates. Introduction With the mapping of the human proteome antibodies, called PLA probes, each with a unique the understanding of protein interactions and short DNA strand (PLUS and MINUS) attached to localization in mammalian cells grows ever it, bind to the primary antibodies. When the PLA more complex. The complexity of the proteome probes are in close proximity (<40 nm), the DNA and the understanding of its inner workings strands can interact through a subsequent addition create a need for more precise and accurate of two other circle-forming DNA oligonucleotides. ways of detecting targets. Besides general After joining of the two added oligonucleotides detection and localization of a protein, it is by enzymatic ligation, they are amplified via rolling necessary to investigate its interaction partners, circle amplification using a polymerase. After the Key Words: posttranslational modifications or its role in amplification reaction (approx 700x) of the DNA protein complexes in order to understand the circle has occurred, labeled complementary Immunofluorescence complete functional role a particular protein plays. -

Application of Proximity Ligation Assay for Multidirectional Studies
To My Family “We must have perseverance and above all confidence in ourselves. We must believe that we are gifted for something and that this thing must be attained.” ― Maria Skłodowska-Curie List of Papers This thesis is based on the following papers, which are referred to in the text by their Roman numerals. I Zieba A, Pardali K, Söderberg O, Lindbom L, Nyström E, Moustakas A, Heldin C-H and Landegren U. Intercellular varia- tion in signaling through the TGF-β pathway and its relation to cell density and cell cycle phase. Mol Cell Proteomics. 2012 Mar 22. [Epub ahead of print] II Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Söder- berg O, Heldin C-H, Landegren U, ten Dijke P and van Dam H. Specific interactions between Smad proteins and AP1 compo- nents determine TGFβ–induced breast cancer cell invasion. Submitted. III Pinidiyaarachchi A, Zieba A, Allalou A, Pardali K, Wählby C. A detailed analysis of 3D subcellular signal localization. Cy- tometry A. 2009 Apr;75(4):319-28. IV Zieba A, Wählby C, Hjelm F, Jordan L, Berg J, Landegren U, Pardali K . Bright-field microscopy visualization of proteins and protein complexes by in situ proximity ligation with perox- idase detection. Clin Chem. 2010 Jan;56(1):99-110. Reprints were made with permission from the respective publishers. Other publications by the author V Raja E, Zieba A, Morén A, Voytyuk I, Söderberg O, Heldin C-H and Moustakas A. The tumour suppressor kinase LKB1 negatively regu- lates bone morphogenetic protein signalling. Submitted. VI Dahl M, Lönn P, Vanlandewijck, M, Zieba A, Hottiger M, Söder- berg O, Heldin C-H and Moustakas A.