Whole Exome and Whole Genome Sequencing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MOLECULAR GENETICIST UBC's Centre for Applied Neurogenetics, Within the Faculty of Medicine, Division of Neurology and Departm
MOLECULAR GENETICIST UBC’s Centre for Applied Neurogenetics, within the Faculty of Medicine, Division of Neurology and Department of Medical Genetics, is seeking a talented and highly motivated Postdoctoral Fellow/Research scientist with a strong background in human genetics. The post- holder will join a multi-discipline team and contribute significantly to identify genetic variability that either causes or contributes to the onset of neurologic and neurodegenerative disease. His/her role will involve working closely with bioinformatician, scientific and medical staff to elucidate the genetic architecture of these diseases (particularly Parkinson’s disease) using state- of-the-art approaches, which range from classical linkage, genome-wide genotyping, through next-generation sequencing, mainly exome and other targeted sequencing experiments (Farrer M. Nat. Rev. Genet. 2006; Farrer M. et al., Nat. Genet. 2008; Vilarino-Guell C. et al., Am. J. Hum. Genet. 2011). Results are used for diagnostic and therapeutic development in partnership with other academic groups and the Pharmaceutical industry (Lewis J. et al., Mol. Neurodegeneration 2008; Melrose H et al., Neurobio Dis. 2010). He/she may also participate in the supervision of interns and graduate students, as well as grant writing. The successful candidate will hold a Ph.D. specializing in human molecular genetics, with an aptitude for molecular biology, statistical genetics and/or bioinformatics. Must have experience with Sanger sequencing, Sequenom, TaqMan and microsatellite genotyping, and strong interest in NGS applied to human disease is desired. He/she will have demonstrated research acumen and have a track record of successful publications, ideally in neurologic and/or neurodegenerative disease. For its beauty and amenities Vancouver is consistently ranked within the top 5 cities to live in the world. -

AMP Molecular Genetic Pathology Recommended Review Books
ASSOCIATION FOR MOLECULAR PATHOLOGY Education. Innovation & Improved Patient Care. Advocacy. 6120 Executive Blvd. Suite 700, Rockville, MD 20852 Tel: 301-634-7939 | Fax: 301-634-7995 | [email protected] | www.amp.org AMP Molecular Genetic Pathology : RECOMMENDED REVIEW BOOKS Updated: March, 2021 The following books are suggested for those preparing for the Molecular Genetic Pathology board exam administered by the American Board of Medical Genetics and the American Board of Pathology. This list represents the recommendations of the MGP Review Course faculty and the AMP Training and Education Committee; endorsement by the ABMG or the ABP is not implied. GENERAL MOLECULAR PATHOLOGY Thompson & Thompson Genetics in Medicine, 8th edition Nussbaum RL et al., eds. Elsevier, 2015 Molecular Pathology: The Molecular Basis of Human Diseases 2nd edition Coleman WB and Tsongalis GJ, eds. Elsevier, 2018 Diagnostic Molecular Pathology: A Guide to Applied Molecular Testing Coleman WB and Tsongalis GJ, eds. Elsevier, 2016 Molecular Basis of Human Cancer, 2nd edition Coleman WB and Tsongalis GJ, eds. Elsevier, 2017 Molecular Diagnostics: Fundamentals, Methods, and Clinical Applications 3rd edition Buckingham L. ASCP, 2021 Genomic Medicine: A Practical Guide Tafe LJ and Arcila ME, eds. Springer, 2020 CYTOGENETICS Gardner and Sutherland’s Chromosome Abnormalities and Genetic Counseling, 5th edition McKinlay Gardner RJ, Amor DJ. Oxford University Press, 2018 The Principles of Clinical Cytogenetics, 3rd edition Gersen SL and Keagle MB, eds. Human Press, 2013 Human Chromosomes, 4th edition Miller OJ and Therman E, eds. Springer Press, 2000 Vance GH, Khan WA. Utility of Fluorescence In Situ Hybridization in Clinical and Research Applications. Advances in Molecular Pathology 2020; 3:65-75. -

Genetics and Other Human Modification Technologies: Sensible International Regulation Or a New Kind of Arms Race?
GENETICS AND OTHER HUMAN MODIFICATION TECHNOLOGIES: SENSIBLE INTERNATIONAL REGULATION OR A NEW KIND OF ARMS RACE? HEARING BEFORE THE SUBCOMMITTEE ON TERRORISM, NONPROLIFERATION, AND TRADE OF THE COMMITTEE ON FOREIGN AFFAIRS HOUSE OF REPRESENTATIVES ONE HUNDRED TENTH CONGRESS SECOND SESSION JUNE 19, 2008 Serial No. 110–201 Printed for the use of the Committee on Foreign Affairs ( Available via the World Wide Web: http://www.foreignaffairs.house.gov/ U.S. GOVERNMENT PRINTING OFFICE 43–068PDF WASHINGTON : 2008 For sale by the Superintendent of Documents, U.S. Government Printing Office Internet: bookstore.gpo.gov Phone: toll free (866) 512–1800; DC area (202) 512–1800 Fax: (202) 512–2104 Mail: Stop IDCC, Washington, DC 20402–0001 COMMITTEE ON FOREIGN AFFAIRS HOWARD L. BERMAN, California, Chairman GARY L. ACKERMAN, New York ILEANA ROS-LEHTINEN, Florida ENI F.H. FALEOMAVAEGA, American CHRISTOPHER H. SMITH, New Jersey Samoa DAN BURTON, Indiana DONALD M. PAYNE, New Jersey ELTON GALLEGLY, California BRAD SHERMAN, California DANA ROHRABACHER, California ROBERT WEXLER, Florida DONALD A. MANZULLO, Illinois ELIOT L. ENGEL, New York EDWARD R. ROYCE, California BILL DELAHUNT, Massachusetts STEVE CHABOT, Ohio GREGORY W. MEEKS, New York THOMAS G. TANCREDO, Colorado DIANE E. WATSON, California RON PAUL, Texas ADAM SMITH, Washington JEFF FLAKE, Arizona RUSS CARNAHAN, Missouri MIKE PENCE, Indiana JOHN S. TANNER, Tennessee JOE WILSON, South Carolina GENE GREEN, Texas JOHN BOOZMAN, Arkansas LYNN C. WOOLSEY, California J. GRESHAM BARRETT, South Carolina SHEILA JACKSON LEE, Texas CONNIE MACK, Florida RUBE´ N HINOJOSA, Texas JEFF FORTENBERRY, Nebraska JOSEPH CROWLEY, New York MICHAEL T. MCCAUL, Texas DAVID WU, Oregon TED POE, Texas BRAD MILLER, North Carolina BOB INGLIS, South Carolina LINDA T. -

Whole Exome and Whole Genome Sequencing – Oxford Clinical Policy
UnitedHealthcare® Oxford Clinical Policy Whole Exome and Whole Genome Sequencing Policy Number: LABORATORY 024.11 T2 Effective Date: October 1, 2021 Instructions for Use Table of Contents Page Related Policies Coverage Rationale ....................................................................... 1 Chromosome Microarray Testing (Non-Oncology Documentation Requirements ...................................................... 2 Conditions) Definitions ...................................................................................... 2 Molecular Oncology Testing for Cancer Diagnosis, Prior Authorization Requirements ................................................ 3 Prognosis, and Treatment Decisions Applicable Codes .......................................................................... 3 • Preimplantation Genetic Testing Description of Services ................................................................. 4 Clinical Evidence ........................................................................... 4 U.S. Food and Drug Administration ........................................... 22 References ................................................................................... 22 Policy History/Revision Information ........................................... 26 Instructions for Use ..................................................................... 27 Coverage Rationale Whole Exome Sequencing (WES) Whole Exome Sequencing (WES) is proven and Medically Necessary for the following: • Diagnosing or evaluating a genetic disorder -

Whole Exome Sequencing Faqs
WHOLE EXOME SEQUENCING (WES) Whole Exome Sequencing (WES) is generally ordered when a patient’s medical history and physical exam strongly suggest that there is an underlying genetic etiology. In some cases, the patient may have had an extensive evaluation consisting of multiple genetic tests, without identifying an etiology. In other cases, a physician may opt to order one of the Whole Exome Sequencing tests early in the patient’s evaluation in an effort to expedite a possible diagnosis and reduce costs incurred by multiple tests. Whole Exome Sequencing is a highly complex test that is newly developed for the identification of changes in a patient’s DNA that are causative or related to their medical concerns. In contrast to current sequencing tests that analyze one gene or small groups of related genes at a time, Whole Exome Sequencing analyzes the exons or coding regions of thousands of genes simultaneously using next-generation sequencing techniques. The exome refers to the portion of the human genome that contains functionally important sequences of DNA that direct the body to make proteins essential for the body to function properly. These regions of DNA are referred to as exons. There are approximately 180,000 exons in the human genome which represents about 3% of the genome. These 180,000 exons are arranged in about 22,000 genes. It is known that many of the errors that occur in DNA sequences that then lead to genetic disorders are located in the exons. Therefore, sequencing of the exome is thought to be an efficient method of analyzing a patient’s DNA to discover the genetic cause of diseases or disabilities. -

University of Wolverhampton
Course Specification Published Date: 14-Sep-2020 Produced By: Laura Clode Status: Validated Core Information Awarding Body / Institution: University of Wolverhampton School / Institute: Wolverhampton School of Sciences Course Code(s): BM021K23UV Sandwich 4 Years UCAS Code: B991 Course Title: BSc (Hons) Biomedical Science with Sandwich Placement Hierarchy of Awards: Bachelor of Science with Honours Biomedical Science, having satisfactorily completed a sandwich placement Bachelor of Science with Honours Biomedical Science, having satisfactorily completed a sandwich placement Bachelor of Science Medical Laboratory Science, having satisfactorily completed a sandwich placement Bachelor of Science Biomedical Science, having satisfactorily completed a sandwich placement Diploma of Higher Education Medical Laboratory Science Certificate of Higher Education Medical Laboratory Science University Statement of Credit University Statement of Credit Language of Study: English Date of DAG approval: 05/Jun/2018 Last Review: 2017/8 Course Specification valid from: 2010/1 Course Specification valid to: 2023/4 Academic Staff Course Leader: Dr Elizabeth O'Gara Head of Department: Dr Elizabeth O'Gara Course Information Location of Delivery: University of Wolverhampton Category of Partnership: Not delivered in partnership Teaching Institution: University of Wolverhampton Open / Closed Course: This course is open to all suitably qualified candidates. Entry Requirements: Entry requirements are subject to regular review. The entry requirements applicable to a particular academic year will be published on the University website (and externally as appropriate e.g. UCAS 240 UCAS points including a science subject at A-level or equivalent. GCSE English and Maths at grade C or above. Distinctive Features of the Course: This course involves the study of a variety of biomedical science disciplines and takes place at an institution where fellow students are undertaking programmes in other disciplines and vocational courses in a wide variety of medicine-related subjects. -

DNA Sequencing and Sorting: Identifying Genetic Variations
BioMath DNA Sequencing and Sorting: Identifying Genetic Variations Student Edition Funded by the National Science Foundation, Proposal No. ESI-06-28091 This material was prepared with the support of the National Science Foundation. However, any opinions, findings, conclusions, and/or recommendations herein are those of the authors and do not necessarily reflect the views of the NSF. At the time of publishing, all included URLs were checked and active. We make every effort to make sure all links stay active, but we cannot make any guaranties that they will remain so. If you find a URL that is inactive, please inform us at [email protected]. DIMACS Published by COMAP, Inc. in conjunction with DIMACS, Rutgers University. ©2015 COMAP, Inc. Printed in the U.S.A. COMAP, Inc. 175 Middlesex Turnpike, Suite 3B Bedford, MA 01730 www.comap.com ISBN: 1 933223 71 5 Front Cover Photograph: EPA GULF BREEZE LABORATORY, PATHO-BIOLOGY LAB. LINDA SHARP ASSISTANT This work is in the public domain in the United States because it is a work prepared by an officer or employee of the United States Government as part of that person’s official duties. DNA Sequencing and Sorting: Identifying Genetic Variations Overview Each of the cells in your body contains a copy of your genetic inheritance, your DNA which has been passed down to you, one half from your biological mother and one half from your biological father. This DNA determines physical features, like eye color and hair color, and can determine susceptibility to medical conditions like hypertension, heart disease, diabetes, and cancer. -
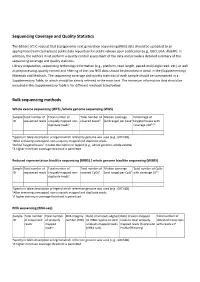
IJC Sequencing Coverage and Quality Statistics
Sequencing Coverage and Quality Statistics The Editors of IJC request that (epi)genomic next generation sequencing (NGS) data should be uploaded to an appropriate (restricted access) public data repository for public release upon publication (e.g., GEO, EGA, dbGAP). In addition, the authors must perform a quality control assessment of the data and provide a detailed summary of the sequencing coverage and quality statistics. Library preparation, sequencing technology information (e.g., platform, read length, paired‐end/single read, etc.) as well as preprocessing, quality control and filtering of the raw NGS data should be described in detail in the (Supplementary) Materials and Methods. The sequencing coverage and quality statistics of each sample should be summarized in a Supplementary Table, to which should be clearly referred in the main text. The minimum information that should be included in this Supplementary Table is for different methods listed below. Bulk sequencing methods Whole exome sequencing (WES) /whole genome sequencing (WGS) Sample Total number of Total number of Total number of Median coverage Percentage of ID sequenced reads uniquely mapped non‐ covered basesb (and range) per baseb targeted bases with duplicate readsa coverage ≥10b,c,d aSpecify in table description or legend which reference genome was used (e.g., GRCh38). bAfter removing unmapped, non‐uniquely mapped and duplicate reads. cDefine “targeted bases” in table description or legend (e.g., whole genome, whole exome). dA higher minimum coverage threshold is permitted. Reduced representation bisulfite sequencing (RRBS) / whole genome bisulfite sequencing (WGBS) Sample Total number of Total number of Total number of Median coverage Total number of CpGs ID sequenced reads uniquely mapped non‐ covered CpGsb (and range) per CpGb with coverage ≥5b,c duplicate readsa aSpecify in table description or legend which reference genome was used (e.g., GRCh38). -

Molecular Diagnostic Testing for Hematology and Oncology Indications Table of Contents Related Coverage Resources
Effective February 15, 2021 Medical Coverage Policy Effective Date.............................................. 2/15/2021 Next Review Date ..................................... 11/15/2021 Coverage Policy Number ................................... 0520 Molecular Diagnostic Testing for Hematology and Oncology Indications Table of Contents Related Coverage Resources Overview ....................................................................... 2 Genetics Coverage Policy ........................................................... 2 Genetic Testing Collateral File General Criteria for Somatic Pathogenic or Likely Pathogenic Variant Genetic Testing ........................... 2 Tumor Profile/Gene Expression Classifier Testing - ...................................................................... 3 Prostate Cancer Screening and Prognostic Tests ..... 6 Tumor Tissue-Based Molecular Assays for Prostate Cancer .......................................................... 6 Hematologic Cancer and Myeloproliferative and Myelodysplastic Disease ............................................ 7 Occult Neoplasms ....................................................... 8 Solid Tumor Cancers .................................................. 9 Other Tumor Profile Testing ....................................... 9 General Background .................................................... 9 General Criteria for Somatic Mutation Genetic Testing ........................................................................ 9 Tumor Profile/Gene Expression Classifier Testing -

Leave Policy for Residents and Fellows.Pdf
Leave Policy for Residents and Fellows The following policy outlines indications and training modifications to accommodate leave during accredited training for certification in all of the ABMGG specialties. Candidates for certification are required to successfully complete training in one of the following programs: ACGME-accredited Programs: 24 months (2 years) • Medical Genetics and Genomics Residency • Clinical Biochemical Genetics Postdoctoral Fellowship • Laboratory Genetics and Genomics Postdoctoral Fellowship Approved Combined Residency Programs: 48 months (4 years) • Medical Genetics and Genomics/Internal Medicine • Medical Genetics and Genomics/Pediatrics • Medical Genetics and Genomics/Maternal Fetal Medicine • Medical Genetics and Genomics/Reproductive Endocrinology and Infertility Subspecialty Fellowship Programs: 12 months (1 year) • Medical Biochemical Genetics • Molecular Genetic Pathology (cosponsored with American Board of Pathology) General Leave Policy: All candidates are allotted 4 weeks leave per year, which may be accrued or averaged through the duration of training for any indication. All candidates must take at least one week off per year as designated vacation time. Leave time cannot be forfeited to shorten training duration. Parental, Caregiver, and Medical Leave Policy: This policy refers to leave for the following indications: parental leave (the birth and care of a newborn including adopted and foster children for either partner), caregiver leave (care for an immediate (first degree) family member or partner with a serious health condition), or medical leave for significant personal medical needs. For Parental, Caregiver, and Medical leave, the ABMGG will allow up to 6 weeks leave during an academic year to occur once during the training program. For trainees who take indicated leave, they must still take at least one additional week for vacation during the same training year. -

The Bio Revolution: Innovations Transforming and Our Societies, Economies, Lives
The Bio Revolution: Innovations transforming economies, societies, and our lives economies, societies, our and transforming Innovations Revolution: Bio The Executive summary The Bio Revolution Innovations transforming economies, societies, and our lives May 2020 McKinsey Global Institute Since its founding in 1990, the McKinsey Global Institute (MGI) has sought to develop a deeper understanding of the evolving global economy. As the business and economics research arm of McKinsey & Company, MGI aims to help leaders in the commercial, public, and social sectors understand trends and forces shaping the global economy. MGI research combines the disciplines of economics and management, employing the analytical tools of economics with the insights of business leaders. Our “micro-to-macro” methodology examines microeconomic industry trends to better understand the broad macroeconomic forces affecting business strategy and public policy. MGI’s in-depth reports have covered more than 20 countries and 30 industries. Current research focuses on six themes: productivity and growth, natural resources, labor markets, the evolution of global financial markets, the economic impact of technology and innovation, and urbanization. Recent reports have assessed the digital economy, the impact of AI and automation on employment, physical climate risk, income inequal ity, the productivity puzzle, the economic benefits of tackling gender inequality, a new era of global competition, Chinese innovation, and digital and financial globalization. MGI is led by three McKinsey & Company senior partners: co-chairs James Manyika and Sven Smit, and director Jonathan Woetzel. Michael Chui, Susan Lund, Anu Madgavkar, Jan Mischke, Sree Ramaswamy, Jaana Remes, Jeongmin Seong, and Tilman Tacke are MGI partners, and Mekala Krishnan is an MGI senior fellow. -

Genomic Sequencing of Infectious Pathogens
Science, Technology Assessment, MARCH 2021 and Analytics WHY THIS MATTERS Genomic sequencing reveals the genetic code of an infectious disease pathogen, such as SARS-CoV-2, the SCIENCE & TECH SPOTLIGHT: virus that causes COVID-19. Newer, faster, and less costly sequencing can now be used to more quickly track GENOMIC SEQUENCING OF transmission, detect new variants, and develop vaccines and other countermeasures. However, challenges such INFECTIOUS PATHOGENS as high startup costs and privacy concerns remain. /// THE TECHNOLOGY How mature is it? First-generation sequencing is often used to confirm results of NGS. NGS is relatively new to the public health field, but is used What is it? Genomic sequencing technologies decode a pathogen’s to augment surveillance (i.e., data collection and analysis) for SARS- genetic material by identifying the order of chemical “letters” of its DNA CoV-2 in the U.S. and overseas. New technologies are allowing greater (or RNA, its chemical equivalent in some viruses). Each of four letters access to sequencing capabilities by making NGS portable, faster, and represents a chemical unit called a base. The sequence of the bases can more affordable (the cost of one sequence run is now one-millionth of reveal useful information for combatting disease. For example, sequencing what it was two decades ago). Genomic sequencing technologies enable of SARS-CoV-2 is used to track the spread of various strains, known as many different areas of infectious disease study. For example, they enable variants (see fig. 1). genomic epidemiology—the science of using pathogen genomic data to determine the distribution and spread of an infectious disease in a group How does it work? Technologies for sequencing genomes of pathogens of people or animals, and the application of this information to respond to use chemicals to break up the DNA or RNA into small fragments.