Outstanding Questions in the Study of Archaic Hominin Admixture
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomics and Its Impact on Science and Society: the Human Genome Project and Beyond
DOE/SC-0083 Genomics and Its Impact on Science and Society The Human Genome Project and Beyond U.S. Department of Energy Genome Research Programs: genomics.energy.gov A Primer ells are the fundamental working units of every living system. All the instructions Cneeded to direct their activities are contained within the chemical DNA (deoxyribonucleic acid). DNA from all organisms is made up of the same chemical and physical components. The DNA sequence is the particular side-by-side arrangement of bases along the DNA strand (e.g., ATTCCGGA). This order spells out the exact instruc- tions required to create a particular organism with protein complex its own unique traits. The genome is an organism’s complete set of DNA. Genomes vary widely in size: The smallest known genome for a free-living organism (a bac- terium) contains about 600,000 DNA base pairs, while human and mouse genomes have some From Genes to Proteins 3 billion (see p. 3). Except for mature red blood cells, all human cells contain a complete genome. Although genes get a lot of attention, the proteins DNA in each human cell is packaged into 46 chro- perform most life functions and even comprise the mosomes arranged into 23 pairs. Each chromosome is majority of cellular structures. Proteins are large, complex a physically separate molecule of DNA that ranges in molecules made up of chains of small chemical com- length from about 50 million to 250 million base pairs. pounds called amino acids. Chemical properties that A few types of major chromosomal abnormalities, distinguish the 20 different amino acids cause the including missing or extra copies or gross breaks and protein chains to fold up into specific three-dimensional rejoinings (translocations), can be detected by micro- structures that define their particular functions in the cell. -

From the Human Genome Project to Genomic Medicine a Journey to Advance Human Health
From the Human Genome Project to Genomic Medicine A Journey to Advance Human Health Eric Green, M.D., Ph.D. Director, NHGRI The Origin of “Genomics”: 1987 Genomics (1987) “For the newly developing discipline of [genome] mapping/sequencing (including the analysis of the information), we have adopted the term GENOMICS… ‘The Genome Institute’ Office for Human Genome Research 1988-1989 National Center for Human Genome Research 1989-1997 National Human Genome Research Institute 1997-present NHGRI: Circa 1990-2003 Human Genome Project NHGRI Today: Characteristic Features . Relatively young (~28 years) . Relatively small (~1.7% of NIH) . Unusual historical origins (think ‘Human Genome Project’) . Emphasis on ‘Team Science’ (think managed ‘consortia’) . Rapidly disseminating footprint (think ‘genomics’) . Novel societal/bioethics research component (think ‘ELSI’) . Over-achievers for trans-NIH initiatives (think ‘Common Fund’) . Vibrant (and large) Intramural Research Program A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project Genomic Medicine An emerging medical discipline that involves using genomic information about an individual as part of their clinical care (e.g., for diagnostic or therapeutic decision- making) and the other implications of that clinical use The Path to Genomic Medicine ? Human Realization of Genome Genomic Project Medicine Nature Nature Base Pairs to Bedside 2003 Heli201x to 1Health A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project -

The Economic Impact and Functional Applications of Human Genetics and Genomics
The Economic Impact and Functional Applications of Human Genetics and Genomics Commissioned by the American Society of Human Genetics Produced by TEConomy Partners, LLC. Report Authors: Simon Tripp and Martin Grueber May 2021 TEConomy Partners, LLC (TEConomy) endeavors at all times to produce work of the highest quality, consistent with our contract commitments. However, because of the research and/or experimental nature of this work, the client undertakes the sole responsibility for the consequence of any use or misuse of, or inability to use, any information or result obtained from TEConomy, and TEConomy, its partners, or employees have no legal liability for the accuracy, adequacy, or efficacy thereof. Acknowledgements ASHG and the project authors wish to thank the following organizations for their generous support of this study. Invitae Corporation, San Francisco, CA Regeneron Pharmaceuticals, Inc., Tarrytown, NY The project authors express their sincere appreciation to the following indi- viduals who provided their advice and input to this project. ASHG Government and Public Advocacy Committee Lynn B. Jorde, PhD ASHG Government and Public Advocacy Committee (GPAC) Chair, President (2011) Professor and Chair of Human Genetics George and Dolores Eccles Institute of Human Genetics University of Utah School of Medicine Katrina Goddard, PhD ASHG GPAC Incoming Chair, Board of Directors (2018-2020) Distinguished Investigator, Associate Director, Science Programs Kaiser Permanente Northwest Melinda Aldrich, PhD, MPH Associate Professor, Department of Medicine, Division of Genetic Medicine Vanderbilt University Medical Center Wendy Chung, MD, PhD Professor of Pediatrics in Medicine and Director, Clinical Cancer Genetics Columbia University Mira Irons, MD Chief Health and Science Officer American Medical Association Peng Jin, PhD Professor and Chair, Department of Human Genetics Emory University Allison McCague, PhD Science Policy Analyst, Policy and Program Analysis Branch National Human Genome Research Institute Rebecca Meyer-Schuman, MS Human Genetics Ph.D. -

Evolutionary Genetics
Evolutionary Genetics Ruben C. Arslan & Lars Penke Institute of Psychology Georg August University Göttingen Forthcoming in D. M. Buss (Ed.), Handbook of Evolutionary Psychology (2nd ed.). New York: Wiley. September 17, 2014 Corresponding author: Ruben C. Arslan Georg August University Göttingen Biological Personality Psychology and Psychological Assessment Georg Elias Müller Institute of Psychology Goßlerstr. 14 37073 Göttingen, Germany Tel.: +49 551 3920704 Email: [email protected] 1 Introduction When Charles Darwin developed the theory of evolution, he knew nothing about genetics. Hence, one of its biggest weaknesses was that Darwin had to base it on crude ideas of inheritance. Around the same time, Gregor Mendel discovered the laws of inheritance, but the scientific community initially failed to appreciate his work’s importance. It was only in the 1930’s that Dobzhansky, Fisher, Haldane, Wright, Mayr and others unified genetics and the theory of evolution in the ‘modern synthesis’. Still, the modern synthesis was built on a basic understanding of genetics, with genes merely being particulate inherited information. The basics of molecular genetics, like the structure of DNA, were not discovered until the 1950’s. When modern evolutionary psychology emerged from ethology and sociobiology in the late 1980’s, it had a strong emphasis on human universals, borne from both the assumption that complex adaptations are monomorphic (or sexually dimorphic) and have to go back to at least the last common ancestor of all humans, and the methodological proximity to experimental cognitive psychology, which tends to treat individual differences as statistical noise. As a consequence, genetic differences between people were marginalized in evolutionary psychology (Tooby & Cosmides, 1990). -

Genetics As Explanation: Limits to the Human Genome Project’ (2006, 2009)
els a0026653.tex V2 - 08/29/2016 4:00 P.M. Page 1 Version 3 a0026653 Genetics as Explanation: Introductory article Article Contents Limits to the Human • Definitions • Metaphors and Programs Genome Project • Metaphors and Expectations • Genome is Not a Simple Program Irun R Cohen, The Weizmann Institute of Science, Rehovot, Israel • Microbes, Symbiosis and the Holobiont Henri Atlan, Ecole des Hautes Etudes en Sciences Sociales, Paris, France and Hadas- • Stem Cells Express Multiple Genes • sah University Hospital, Jerusalem, Israel Meaning: Line or Loop? AU:1 • Self-Organisation and Program Sol Efroni, Bar-Ilan University, Ramat Gan, Israel • Complexity, Reduction and Emergence • Evolving Genomes AU:2 Based in part on the previous versions of this eLS article ‘Genetics as Explanation: Limits to the Human Genome Project’ (2006, 2009). • The Environment and the Genome • Language Metaphor • Tool and Toolbox Metaphors • Conclusion Online posting date: 14th October 2016 Living organisms are composed of cells and all biological evolution, single organisms, populations of organisms, living cells contain a genome, the organism’s stock species, cells and molecules, heredity, embryonic development, of deoxyribonucleic acid (DNA). The role of the health and disease and life management. These are quite diverse genome has been likened to a computer program subjects, and the people who study them would seem to use the term gene in distinctly different ways. But genetics as a whole that encodes the organism’s development and its is organised by a single unifying principle, the deoxyribonucleic subsequent response to the environment. Thus, the acid (DNA) code; all would agree that the information borne by organism and its fate can be explained by genetics, a gene is linked to particular sequences of DNA. -

The Human Genome Project: Frequently Asked Questions
The Human Genome Project: Frequently Asked Questions On April 14, 2003, the National Human Genome Research Institute (NHGRI), the Department of Energy (DOE) and their partners in the International Human Genome Sequencing Consortium announced the successful completion of the Human Genome Project. What is a genome? A genome is an organism's complete set of deoxyribonucleic acid (DNA), a chemical compound that contains the genetic instructions needed to develop and direct the biological activities of every organism. DNA molecules are made of two twisting, paired strands. Each strand is made of four chemical units, called nucleotide bases. The bases are adenine (A), thymine (T), guanine (G) and cytosine (C). Bases on opposite strands pair specifically: an A always pairs with a T, and a C always with a G. The human genome contains approximately 3 billion of these base pairs, which reside in the 23 pairs of chromosomes within the nucleus of all our cells. The Human Genome’s Project’s principal task was determining the order or sequence of those As, Ts, Cs, and Gs. What is sequencing and how do you sequence a genome? Sequencing means determining the exact order of the base pairs in a segment of DNA. Human chromosomes range in size from about 50 million to 300 million base pairs. One complete set of genomic instructions is about 3 billion base pairs. Specialized machines called DNA sequencers – the technologies have rapidly evolved over the last decade – use techniques of biochemistry to automatically determine the base pair order and deposit the read out in a computer. -

Genomics and Human Identity Grades 7-12
Genomics and Human Identity Grades 7-12 Lesson 1 Inspired by the museum exhibit Genome: Unlocking Life’s Code unlockinglifescode.org Smithsonian National Museum of Natural History Genomics and Human Identity: Lesson 1 Genomics and Human Identity: Lesson Table of Contents Table of Contents.................................................................................................................2 About the Genomics and Human Identity lessons ..................................................3 Introduction to Lesson 1 – Alike and Different .........................................................3 Lesson Components ............................................................................................................4 Time .........................................................................................................................................4 Key Concepts .........................................................................................................................4 Learning Objectives ............................................................................................................4 Next Generation Science Standards .............................................................................4 Prerequisite Knowledge ....................................................................................................5 Materials and Handouts ....................................................................................................5 Preparation ............................................................................................................................6 -

Initial Upper Palaeolithic Humans in Europe Had Recent Neanderthal Ancestry
Article Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry https://doi.org/10.1038/s41586-021-03335-3 Mateja Hajdinjak1,2 ✉, Fabrizio Mafessoni1, Laurits Skov1, Benjamin Vernot1, Alexander Hübner1,3, Qiaomei Fu4, Elena Essel1, Sarah Nagel1, Birgit Nickel1, Julia Richter1, Received: 7 July 2020 Oana Teodora Moldovan5,6, Silviu Constantin7,8, Elena Endarova9, Nikolay Zahariev10, Accepted: 5 February 2021 Rosen Spasov10, Frido Welker11,12, Geoff M. Smith11, Virginie Sinet-Mathiot11, Lindsey Paskulin13, Helen Fewlass11, Sahra Talamo11,14, Zeljko Rezek11,15, Svoboda Sirakova16, Nikolay Sirakov16, Published online: 7 April 2021 Shannon P. McPherron11, Tsenka Tsanova11, Jean-Jacques Hublin11,17, Benjamin M. Peter1, Open access Matthias Meyer1, Pontus Skoglund2, Janet Kelso1 & Svante Pääbo1 ✉ Check for updates Modern humans appeared in Europe by at least 45,000 years ago1–5, but the extent of their interactions with Neanderthals, who disappeared by about 40,000 years ago6, and their relationship to the broader expansion of modern humans outside Africa are poorly understood. Here we present genome-wide data from three individuals dated to between 45,930 and 42,580 years ago from Bacho Kiro Cave, Bulgaria1,2. They are the earliest Late Pleistocene modern humans known to have been recovered in Europe so far, and were found in association with an Initial Upper Palaeolithic artefact assemblage. Unlike two previously studied individuals of similar ages from Romania7 and Siberia8 who did not contribute detectably to later populations, these individuals are more closely related to present-day and ancient populations in East Asia and the Americas than to later west Eurasian populations. This indicates that they belonged to a modern human migration into Europe that was not previously known from the genetic record, and provides evidence that there was at least some continuity between the earliest modern humans in Europe and later people in Eurasia. -

Human Chromosome-Specific Cdna Libraries: New Tools for Gene Identification and Genome Annotation
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH Human Chromosome-specific cDNA Libraries: New Tools for Gene Identification and Genome Annotation Richard G. Del Mastro, 1'2 Luping Wang, ~'2 Andrew D. Simmons, Teresa D. Gallardo, 1 Gregory A. Clines, ~ Jennifer A. Ashley, 1 Cynthia J. Hilliard, 3 John J. Wasmuth, 3 John D. McPherson, 3 and Michael Lovett ~'4 1Department of Biochemistry and the McDermott Center for Human Growth and Development, The University of Texas Southwestern Medical Center, Dallas, Texas 75235-8591; 3Department of Biological Chemistry and the Human Genome Center, University of California, Irvine, California 9271 7 To date, only a small percentage of human genes have been cloned and mapped. To facilitate more rapid gene mapping and disease gene isolation, chromosome S-specific cDNA libraries have been constructed from five sources. DNA sequencing and regional mapping of 205 unique cDNAs indicates that 25 are from known chromosome S genes and 138 are from new chromosome S genes (a frequency of 79.5%}. Sequence complexity estimates indicate that each library contains -20% of the -SO00 genes that are believed to reside on chromosome 5. This study more than doubles the number of genes mapped to chromosome S and describes an important new tool for disease gene isolation. A detailed map of expressed sequences within the pressed Sequence Tags (eSTs)] (Adams et al. 1991, human genome would provide an indispensable 1992, 1993a,b; Khan et al. 1991; Wilcox et al. resource for isolating disease genes, and would 1991; Okubo et al. -

Oldest-Known Human Genome Sequenced
IN FOCUS NEWS US$750 million to send military personnel PALAEOANTHROPOLOGY and other resources to the outbreak zone. Funding for many science agencies did rise slightly overall in the 2014 fiscal year after the mandatory sequestration in 2013. Oldest-known human The National Oceanic and Atmospheric Administration’s budget actually rose by roughly $575 million in 2014, to $5.3 billion. genome sequenced But the National Institutes of Health (NIH), the world’s largest biomedical-research funder, received $29.9 billion, less than its DNA shows a group of modern humans roamed across Asia. pre-sequester budget of $30.7 billion. “Everybody in Congress knows that BY EWEN CALLAWAY world, but later went extinct, Viola says. science is important,” says Congressman The most intriguing clue about his origin Rush Holt (Democrat, New Jersey), “but 45,000-year-old leg bone from Siberia is that about 2% of his genome comes from they don’t have much appreciation of what it has yielded the oldest genome sequence Neanderthals. This is roughly the same level takes to sustain it.” (Physicist Holt, one of the for Homo sapiens on record — reveal- that lurks in the genomes of all of today’s non- few scientists in US public office, famously Aing a mysterious population that may once have Africans, owing to ancient trysts between their gives out bumper stickers that read, “My spanned northern Asia. The DNA sequence ancestors and Neanderthals. The Ust’-Ishim Congressman IS a Rocket Scientist!”) from a male hunter-gatherer also offers tantali man probably got his Neanderthal DNA The NIH cuts have put the squeeze on zing clues about modern humans’ journey from from these same matings, which, past studies many research institutions. -
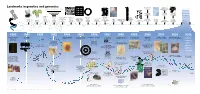
Landmarks in Genetics and Genomics
0 ++- ---+ BC PR • --- -+-t--.--+------ o,ou 1J.T33.? n.e -- -- - 4 Landmarks in genetics and genomics !!!!! U C AG +++j/GA T C US National Research U Phe Tyr Cys International Nucleotide Ser C Council issues report on U Leu stop stop A Sequence Database - stop Trp - G Mapping and Sequencing the U Consortium formed Leu Pro His Arg C Human Genome 0 5' C Gln A 3' G Human U 5' lle Asn Ser STS Markers Thr C A Met Lys Arg A G Muscular-dystrophy Development New strands Sequence-tagged Asp U of yeast artificial Genome G Val Ala Gly C gene identified sites (STS) mapping Glu A chromosome (YAC) G by positional cloning concept established cloning Archibald Garrod Project formulates Alfred Henry Oswald Avery, Colin MacLeod James Watson and Stanley Cohen and Frederick Sanger, First public Desired fragment the concept Sturtevant and Maclyn McCarty Francis Crick Marshall Nirenberg, Herbert Boyer The Belmont Report Allan Maxam discussion strands Gregor Mendel of human makes demonstrate that DNA describe the Har Gobind Khorana and develop on the use of and Walter Gilbert GenBank First human disease gene — for of sequencing First automated First-generation Cystic-fibrosis discovers Rediscovery of inborn errors the first linear is the double-helical Robert Holley determine recombinant human subjects develop DNA-sequencing database Huntington’s disease — is mapped the human The polymerase DNA-sequencing instrument human genetic Human Genome gene identified by laws of genetics Mendel’s work of metabolism map of genes hereditary material structure of DNA the genetic code DNA technology in research is issued methods established with DNA markers genome chain reaction (PCR) is invented developed map developed Organization (HUGO) formed positional cloning 1865 1900 1905 1913 1944 1953 1966 1972 1974 1977 1982 1983 1984 1985 1986 1987 1988 1989 1990 2003 +-+-o-----+---+ 1 I~I "-.■ u ''\. -

Human Genetic Variation and Disease
Resource pack: Human genetic variation and disease Scottish Curriculum links Higher Biology: DNA and the Genome Unit Study of DNA and the genome. The Unit covers the key areas of structure of DNA; replication of DNA; control of gene expression; cellular differentiation; the structure of the genome; mutations; evolution; genomic sequencing. Personal genomics. Higher Human Biology: Human Cells Unit Structure and Replication of DNA, Gene Expression, Genes and Proteins in Health and Disease, Human Genomics. The Unit covers the key areas of structure and replication of DNA; gene expression; genes and proteins in health and disease, human genomics. Expression of genotype and protein production which allows study of mutations and disorders National 5 Biology: Multicellular Organisms Unit Reproduction, variation and inheritance; the need for transport and effects of life-style choices on animal transport and exchange systems. Resource pack contents This resource pack provides information on: Genetic variation/ DNA mutations Genetic basis of disease risk Human population cohort studies and methods Genes and Proteins in Health and Disease Human Genomics Expression of genotype and protein production For more information on obesity and diabetes, mentioned as example diseases here, another resource pack is available: www.sserc.org.uk/images/Biology/Higher_Human/SQA/Diabetes_final.pdf Page 1 Background Many common diseases are influenced by a combination of multiple genes and environmental factors and are therefore referred to as complex diseases. Because they can be caused by both genetic and environmental factors, complex diseases – such as asthma, stroke and diabetes - can be difficult to treat. In 1990, a massive international scientific project called The Human Genome Project was launched to determine the sequence of the building blocks/letters/nucleotides of the human DNA code - the entire human genome.