Searching Scale Space for Activation in PET Images
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Scale Space Multiresolution Analysis of Random Signals
Scale Space Multiresolution Analysis of Random Signals Lasse Holmstr¨om∗, Leena Pasanen Department of Mathematical Sciences, University of Oulu, Finland Reinhard Furrer Institute of Mathematics, University of Zurich,¨ Switzerland Stephan R. Sain National Center for Atmospheric Research, Boulder, Colorado, USA Abstract A method to capture the scale-dependent features in a random signal is pro- posed with the main focus on images and spatial fields defined on a regular grid. A technique based on scale space smoothing is used. However, where the usual scale space analysis approach is to suppress detail by increasing smoothing progressively, the proposed method instead considers differences of smooths at neighboring scales. A random signal can then be represented as a sum of such differences, a kind of a multiresolution analysis, each difference representing de- tails relevant at a particular scale or resolution. Bayesian analysis is used to infer which details are credible and which are just artifacts of random variation. The applicability of the method is demonstrated using noisy digital images as well as global temperature change fields produced by numerical climate prediction models. Keywords: Scale space smoothing, Bayesian methods, Image analysis, Climate research 1. Introduction In signal processing, smoothing is often used to suppress noise. An optimal level of smoothing is then of interest. In scale space analysis of noisy curves and images, instead of a single, in some sense “optimal” level of smoothing, a whole family of smooths is considered and each smooth is thought to provide information about the underlying object at a particular scale or resolution. The ∗Corresponding author ∗∗P.O.Box 3000, 90014 University of Oulu, Finland Tel. -

Image Denoising Using Non Linear Diffusion Tensors
2011 8th International Multi-Conference on Systems, Signals & Devices IMAGE DENOISING USING NON LINEAR DIFFUSION TENSORS Benzarti Faouzi, Hamid Amiri Signal, Image Processing and Pattern Recognition: TSIRF (ENIT) Email: [email protected] , [email protected] ABSTRACT linear models. Many approaches have been proposed to remove the noise effectively while preserving the original Image denoising is an important pre-processing step for image details and features as much as possible. In the past many image analysis and computer vision system. It few years, the use of non linear PDEs methods involving refers to the task of recovering a good estimate of the true anisotropic diffusion has significantly grown and image from a degraded observation without altering and becomes an important tool in contemporary image changing useful structure in the image such as processing. The key idea behind the anisotropic diffusion discontinuities and edges. In this paper, we propose a new is to incorporate an adaptative smoothness constraint in approach for image denoising based on the combination the denoising process. That is, the smooth is encouraged of two non linear diffusion tensors. One allows diffusion in a homogeneous region and discourage across along the orientation of greatest coherences, while the boundaries, in order to preserve the natural edge of the other allows diffusion along orthogonal directions. The image. One of the most successful tools for image idea is to track perfectly the local geometry of the denoising is the Total Variation (TV) model [9] [8][10] degraded image and applying anisotropic diffusion and the anisotropic smoothing model [1] which has since mainly along the preferred structure direction. -

Scale-Space Theory for Multiscale Geometric Image Analysis
Tutorial Scale-Space Theory for Multiscale Geometric Image Analysis Bart M. ter Haar Romeny, PhD Utrecht University, the Netherlands [email protected] Introduction Multiscale image analysis has gained firm ground in computer vision, image processing and models of biological vision. The approaches however have been characterised by a wide variety of techniques, many of them chosen ad hoc. Scale-space theory, as a relatively new field, has been established as a well founded, general and promising multiresolution technique for image structure analysis, both for 2D, 3D and time series. The rather mathematical nature of many of the classical papers in this field has prevented wide acceptance so far. This tutorial will try to bridge that gap by giving a comprehensible and intuitive introduction to this field. We also try, as a mutual inspiration, to relate the computer vision modeling to biological vision modeling. The mathematical rigor is much relaxed for the purpose of giving the broad picture. In appendix A a number of references are given as a good starting point for further reading. The multiscale nature of things In mathematics objects have no scale. We are familiar with the notion of points, that really shrink to zero, lines with zero width. In mathematics are no metrical units involved, as in physics. Neighborhoods, like necessary in the definition of differential operators, are defined as taken into the limit to zero, so we can really speak of local operators. In physics objects live on a range of scales. We need an instrument to do an observation (our eye, a camera) and it is the range that this instrument can see that we call the scale range. -

An Overview of Wavelet Transform Concepts and Applications
An overview of wavelet transform concepts and applications Christopher Liner, University of Houston February 26, 2010 Abstract The continuous wavelet transform utilizing a complex Morlet analyzing wavelet has a close connection to the Fourier transform and is a powerful analysis tool for decomposing broadband wavefield data. A wide range of seismic wavelet applications have been reported over the last three decades, and the free Seismic Unix processing system now contains a code (succwt) based on the work reported here. Introduction The continuous wavelet transform (CWT) is one method of investigating the time-frequency details of data whose spectral content varies with time (non-stationary time series). Moti- vation for the CWT can be found in Goupillaud et al. [12], along with a discussion of its relationship to the Fourier and Gabor transforms. As a brief overview, we note that French geophysicist J. Morlet worked with non- stationary time series in the late 1970's to find an alternative to the short-time Fourier transform (STFT). The STFT was known to have poor localization in both time and fre- quency, although it was a first step beyond the standard Fourier transform in the analysis of such data. Morlet's original wavelet transform idea was developed in collaboration with the- oretical physicist A. Grossmann, whose contributions included an exact inversion formula. A series of fundamental papers flowed from this collaboration [16, 12, 13], and connections were soon recognized between Morlet's wavelet transform and earlier methods, including harmonic analysis, scale-space representations, and conjugated quadrature filters. For fur- ther details, the interested reader is referred to Daubechies' [7] account of the early history of the wavelet transform. -
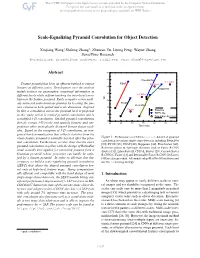
Scale-Equalizing Pyramid Convolution for Object Detection
Scale-Equalizing Pyramid Convolution for Object Detection Xinjiang Wang,* Shilong Zhang∗, Zhuoran Yu, Litong Feng, Wayne Zhang SenseTime Research {wangxinjiang, zhangshilong, yuzhuoran, fenglitong, wayne.zhang}@sensetime.com Abstract 42 41 FreeAnchor C-Faster Feature pyramid has been an efficient method to extract FSAF D-Faster 40 features at different scales. Development over this method Reppoints mainly focuses on aggregating contextual information at 39 different levels while seldom touching the inter-level corre- L-Faster AP lation in the feature pyramid. Early computer vision meth- 38 ods extracted scale-invariant features by locating the fea- FCOS RetinaNet ture extrema in both spatial and scale dimension. Inspired 37 by this, a convolution across the pyramid level is proposed Faster Baseline 36 SEPC-lite in this study, which is termed pyramid convolution and is Two-stage detectors a modified 3-D convolution. Stacked pyramid convolutions 35 directly extract 3-D (scale and spatial) features and out- 60 65 70 75 80 85 90 performs other meticulously designed feature fusion mod- Time (ms) ules. Based on the viewpoint of 3-D convolution, an inte- grated batch normalization that collects statistics from the whole feature pyramid is naturally inserted after the pyra- Figure 1: Performance on COCO-minival dataset of pyramid mid convolution. Furthermore, we also show that the naive convolution in various single-stage detectors including RetinaNet [20], FCOS [38], FSAF [48], Reppoints [44], FreeAnchor [46]. pyramid convolution, together with the design of RetinaNet Reference points of two-stage detectors such as Faster R-CNN head, actually best applies for extracting features from a (Faster) [31], Libra Faster R-CNN (L-Faster) [29], Cascade Faster Gaussian pyramid, whose properties can hardly be satis- R-CNN (C-Faster) [1] and Deformable Faster R-CNN (D-Faster) fied by a feature pyramid. -

Scale Normalized Image Pyramids with Autofocus for Object Detection
1 Scale Normalized Image Pyramids with AutoFocus for Object Detection Bharat Singh, Mahyar Najibi, Abhishek Sharma and Larry S. Davis Abstract—We present an efficient foveal framework to perform object detection. A scale normalized image pyramid (SNIP) is generated that, like human vision, only attends to objects within a fixed size range at different scales. Such a restriction of objects’ size during training affords better learning of object-sensitive filters, and therefore, results in better accuracy. However, the use of an image pyramid increases the computational cost. Hence, we propose an efficient spatial sub-sampling scheme which only operates on fixed-size sub-regions likely to contain objects (as object locations are known during training). The resulting approach, referred to as Scale Normalized Image Pyramid with Efficient Resampling or SNIPER, yields up to 3× speed-up during training. Unfortunately, as object locations are unknown during inference, the entire image pyramid still needs processing. To this end, we adopt a coarse-to-fine approach, and predict the locations and extent of object-like regions which will be processed in successive scales of the image pyramid. Intuitively, it’s akin to our active human-vision that first skims over the field-of-view to spot interesting regions for further processing and only recognizes objects at the right resolution. The resulting algorithm is referred to as AutoFocus and results in a 2.5-5× speed-up during inference when used with SNIP. Code: https://github.com/mahyarnajibi/SNIPER Index Terms—Object Detection, Image Pyramids , Foveal vision, Scale-Space Theory, Deep-Learning. F 1 INTRODUCTION BJECT-detection is one of the most popular and widely O researched problems in the computer vision community, owing to its application to a myriad of industrial systems, such as autonomous driving, robotics, surveillance, activity-detection, scene-understanding and/or large-scale multimedia analysis. -

Multi-Scale Edge Detection and Image Segmentation
MULTI-SCALE EDGE DETECTION AND IMAGE SEGMENTATION Baris Sumengen, B. S. Manjunath ECE Department, UC, Santa Barbara 93106, Santa Barbara, CA, USA email: {sumengen,manj}@ece.ucsb.edu web: vision.ece.ucsb.edu ABSTRACT In this paper, we propose a novel multi-scale edge detection and vector field design scheme. We show that using multi- scale techniques edge detection and segmentation quality on natural images can be improved significantly. Our ap- proach eliminates the need for explicit scale selection and edge tracking. Our method favors edges that exist at a wide range of scales and localize these edges at finer scales. This work is then extended to multi-scale image segmentation us- (a) (b) (c) (d) ing our anisotropic diffusion scheme. Figure 1: Localized and clean edges using multiple scales. a) 1. INTRODUCTION Original image. b) Edge strengths at spatial scale σ = 1, c) Most edge detection algorithms specify a spatial scale at using scales from σ = 1 to σ = 4. d) at σ = 4. Edges are not which the edges are detected. Typically, edge detectors uti- well localized at σ = 4. lize local operators and the effective area of these local oper- ators define this spatial scale. The spatial scale usually corre- sponds to the level of smoothing of the image, for example, scales and generating a synthesis of these edges. On the the variance of the Gaussian smoothing. At small scales cor- other hand, it is desirable that the multi-scale information responding to finer image details, edge detectors find inten- is integrated to the edge detection at an earlier stage and the sity jumps in small neighborhoods. -

Computer Vision: Edge Detection
Edge Detection Edge detection Convert a 2D image into a set of curves • Extracts salient features of the scene • More compact than pixels Origin of Edges surface normal discontinuity depth discontinuity surface color discontinuity illumination discontinuity Edges are caused by a variety of factors Edge detection How can you tell that a pixel is on an edge? Profiles of image intensity edges Edge detection 1. Detection of short linear edge segments (edgels) 2. Aggregation of edgels into extended edges (maybe parametric description) Edgel detection • Difference operators • Parametric-model matchers Edge is Where Change Occurs Change is measured by derivative in 1D Biggest change, derivative has maximum magnitude Or 2nd derivative is zero. Image gradient The gradient of an image: The gradient points in the direction of most rapid change in intensity The gradient direction is given by: • how does this relate to the direction of the edge? The edge strength is given by the gradient magnitude The discrete gradient How can we differentiate a digital image f[x,y]? • Option 1: reconstruct a continuous image, then take gradient • Option 2: take discrete derivative (finite difference) How would you implement this as a cross-correlation? The Sobel operator Better approximations of the derivatives exist • The Sobel operators below are very commonly used -1 0 1 1 2 1 -2 0 2 0 0 0 -1 0 1 -1 -2 -1 • The standard defn. of the Sobel operator omits the 1/8 term – doesn’t make a difference for edge detection – the 1/8 term is needed to get the right gradient -

Scale-Space and Edge Detection Using Anisotropic Diffusion
IEEE TRANSACTIONS ON PATTERN ANALYSIS AND MACHINE INTELLIGENCE, VOL. 12. NO. 7. JULY 1990 629 Scale-Space and Edge Detection Using Anisotropic Diffusion PIETRO PERONA AND JITENDRA MALIK Abstracf-The scale-space technique introduced by Witkin involves generating coarser resolution images by convolving the original image with a Gaussian kernel. This approach has a major drawback: it is difficult to obtain accurately the locations of the “semantically mean- ingful” edges at coarse scales. In this paper we suggest a new definition of scale-space, and introduce a class of algorithms that realize it using a diffusion process. The diffusion coefficient is chosen to vary spatially in such a way as to encourage intraregion smoothing in preference to interregion smoothing. It is shown that the “no new maxima should be generated at coarse scales” property of conventional scale space is pre- served. As the region boundaries in our approach remain sharp, we obtain a high quality edge detector which successfully exploits global information. Experimental results are shown on a number of images. The algorithm involves elementary, local operations replicated over the Fig. 1. A family of l-D signals I(x, t)obtained by convolving the original image making parallel hardware implementations feasible. one (bottom) with Gaussian kernels whose variance increases from bot- tom to top (adapted from Witkin [21]). Zndex Terms-Adaptive filtering, analog VLSI, edge detection, edge enhancement, nonlinear diffusion, nonlinear filtering, parallel algo- rithm, scale-space. with the initial condition I(x, y, 0) = Zo(x, y), the orig- inal image. 1. INTRODUCTION Koenderink motivates the diffusion equation formula- HE importance of multiscale descriptions of images tion by stating two criteria. -

Scalespaceviz: α-Scale Spaces in Practice
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector IMAGE PROCESSING, ANALYSIS, RECOGNITION, AND UNDERSTANDING ScaleSpaceViz: a-Scale Spaces in Practice¶ F. Kanters, L. Florack, R. Duits, B. Platel, and B. ter Haar Romeny Eindhoven University of Technology Department of Biomedical Engineering Den Dolech 2, Postbus 513 5600 MB Eindhoven, Netherlands e-mail: {F.M.W.Kanters, L.M.J.Florack, R.Duits, B.Platel, B.M.terHaarRomeny}@tue.nl Abstract—Kernels of the so-called α-scale space have the undesirable property of having no closed-form rep- resentation in the spatial domain, despite their simple closed-form expression in the Fourier domain. This obstructs spatial convolution or recursive implementation. For this reason an approximation of the 2D α-kernel in the spatial domain is presented using the well-known Gaussian kernel and the Poisson kernel. Experiments show good results, with maximum relative errors of less than 2.4%. The approximation has been successfully implemented in a program for visualizing α-scale spaces. Some examples of practical applications with scale space feature points using the proposed approximation are given. DOI: 10.1134/S1054661807010129 1. INTRODUCTION Witkin [2] and Koenderink [3] in the 1980s. A more recent introduction to scale space can be found in Scale is an essential parameter in computer vision, monograph by ter Haar Romeny [4]. It is, however, since it is an immediate consequence of the process of shown by Duits [5] that, for reasonable axioms, there observation, of measurements. Observations are always exists a complete class of kernels leading to the so- done by integrating some physical property with a called α-scale spaces, of which the Gaussian scale measurement device—for example, integration (over a space is a special case (α = 1). -

Scale-Space and Edge Detection Using Anisotropic Diffusion
IEEE TRANSACTIONS ON PATTERN ANALYSIS AND MACHINE INTELLIGENCE, VOL. 12. NO. 7. JULY 1990 629 Scale-Space and Edge Detection Using Anisotropic Diffusion PIETRO PERONA AND JITENDRA MALIK Abstracf-The scale-space technique introduced by Witkin involves generating coarser resolution images by convolving the original image with a Gaussian kernel. This approach has a major drawback: it is difficult to obtain accurately the locations of the “semantically mean- ingful” edges at coarse scales. In this paper we suggest a new definition of scale-space, and introduce a class of algorithms that realize it using a diffusion process. The diffusion coefficient is chosen to vary spatially in such a way as to encourage intraregion smoothing in preference to interregion smoothing. It is shown that the “no new maxima should be generated at coarse scales” property of conventional scale space is pre- served. As the region boundaries in our approach remain sharp, we obtain a high quality edge detector which successfully exploits global information. Experimental results are shown on a number of images. The algorithm involves elementary, local operations replicated over the Fig. 1. A family of l-D signals I(x, t)obtained by convolving the original image making parallel hardware implementations feasible. one (bottom) with Gaussian kernels whose variance increases from bot- tom to top (adapted from Witkin [21]). Zndex Terms-Adaptive filtering, analog VLSI, edge detection, edge enhancement, nonlinear diffusion, nonlinear filtering, parallel algo- rithm, scale-space. with the initial condition I(x, y, 0) = Zo(x, y), the orig- inal image. 1. INTRODUCTION Koenderink motivates the diffusion equation formula- HE importance of multiscale descriptions of images tion by stating two criteria. -

Local Descriptors
Local descriptors SIFT (Scale Invariant Feature Transform) descriptor • SIFT keypoints at locaon xy and scale σ have been obtained according to a procedure that guarantees illuminaon and scale invance. By assigning a consistent orientaon, the SIFT keypoint descriptor can be also orientaon invariant. • SIFT descriptor is therefore obtained from the following steps: – Determine locaon and scale by maximizing DoG in scale and in space (i.e. find the blurred image of closest scale) – Sample the points around the keypoint and use the keypoint local orientaon as the dominant gradient direc;on. Use this scale and orientaon to make all further computaons invariant to scale and rotaon (i.e. rotate the gradients and coordinates by the dominant orientaon) – Separate the region around the keypoint into subregions and compute a 8-bin gradient orientaon histogram of each subregion weigthing samples with a σ = 1.5 Gaussian SIFT aggregaon window • In order to derive a descriptor for the keypoint region we could sample intensi;es around the keypoint, but they are sensi;ve to ligh;ng changes and to slight errors in x, y, θ. In order to make keypoint es;mate more reliable, it is usually preferable to use a larger aggregaon window (i.e. the Gaussian kernel size) than the detec;on window. The SIFT descriptor is hence obtained from thresholded image gradients sampled over a 16x16 array of locaons in the neighbourhood of the detected keypoint, using the level of the Gaussian pyramid at which the keypoint was detected. For each 4x4 region samples they are accumulated into a gradient orientaon histogram with 8 sampled orientaons.