Interpretation of Mammogram and Chest Radiograph Reports Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACR–SPR-STR Practice Parameter for the Performance of Chest Radiography
The American College of Radiology, with more than 30,000 members, is the principal organization of radiologists, radiation oncologists, and clinical medical physicists in the United States. The College is a nonprofit professional society whose primary purposes are to advance the science of radiology, improve radiologic services to the patient, study the socioeconomic aspects of the practice of radiology, and encourage continuing education for radiologists, radiation oncologists, medical physicists, and persons practicing in allied professional fields. The American College of Radiology will periodically define new practice parameters and technical standards for radiologic practice to help advance the science of radiology and to improve the quality of service to patients throughout the United States. Existing practice parameters and technical standards will be reviewed for revision or renewal, as appropriate, on their fifth anniversary or sooner, if indicated. Each practice parameter and technical standard, representing a policy statement by the College, has undergone a thorough consensus process in which it has been subjected to extensive review and approval. The practice parameters and technical standards recognize that the safe and effective use of diagnostic and therapeutic radiology requires specific training, skills, and techniques, as described in each document. Reproduction or modification of the published practice parameter and technical standard by those entities not providing these services is not authorized. Revised 2017 (Resolution 2)* ACR–SPR–STR PRACTICE PARAMETER FOR THE PERFORMANCE OF CHEST RADIOGRAPHY PREAMBLE This document is an educational tool designed to assist practitioners in providing appropriate radiologic care for patients. Practice Parameters and Technical Standards are not inflexible rules or requirements of practice and are not intended, nor should they be used, to establish a legal standard of care1. -

VIEW Open Access Ultrasound and Non‑Ultrasound Imaging Techniques in the Assessment of Diaphragmatic Dysfunction Franco A
Laghi Jr. et al. BMC Pulm Med (2021) 21:85 https://doi.org/10.1186/s12890-021-01441-6 REVIEW Open Access Ultrasound and non-ultrasound imaging techniques in the assessment of diaphragmatic dysfunction Franco A. Laghi Jr.1, Marina Saad2 and Hameeda Shaikh3,4* Abstract Diaphragm muscle dysfunction is increasingly recognized as an important element of several diseases including neuromuscular disease, chronic obstructive pulmonary disease and diaphragm dysfunction in critically ill patients. Functional evaluation of the diaphragm is challenging. Use of volitional maneuvers to test the diaphragm can be limited by patient efort. Non-volitional tests such as those using neuromuscular stimulation are technically complex, since the muscle itself is relatively inaccessible. As such, there is a growing interest in using imaging techniques to characterize diaphragm muscle dysfunction. Selecting the appropriate imaging technique for a given clinical sce- nario is a critical step in the evaluation of patients suspected of having diaphragm dysfunction. In this review, we aim to present a detailed analysis of evidence for the use of ultrasound and non-ultrasound imaging techniques in the assessment of diaphragm dysfunction. We highlight the utility of the qualitative information gathered by ultrasound imaging as a means to assess integrity, excursion, thickness, and thickening of the diaphragm. In contrast, quantitative ultrasound analysis of the diaphragm is marred by inherent limitations of this technique, and we provide a detailed examination of these limitations. We evaluate non-ultrasound imaging modalities that apply static techniques (chest radiograph, computerized tomography and magnetic resonance imaging), used to assess muscle position, shape and dimension. We also evaluate non-ultrasound imaging modalities that apply dynamic imaging (fuoroscopy and dynamic magnetic resonance imaging) to assess diaphragm motion. -

Diagnostic Use of Lung Ultrasound Compared to Chest Radiograph for Suspected Pneumonia in a Resource-Limited Setting Yogendra Amatya1, Jordan Rupp2, Frances M
Amatya et al. International Journal of Emergency Medicine (2018) 11:8 International Journal of https://doi.org/10.1186/s12245-018-0170-2 Emergency Medicine ORIGINAL RESEARCH Open Access Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting Yogendra Amatya1, Jordan Rupp2, Frances M. Russell3, Jason Saunders3, Brian Bales2 and Darlene R. House1,3* Abstract Background: Lung ultrasound is an effective tool for diagnosing pneumonia in developed countries. Diagnostic accuracy in resource-limited countries where pneumonia is the leading cause of death is unknown. The objective of this study was to evaluate the sensitivity of bedside lung ultrasound compared to chest X-ray for pneumonia in adults presenting for emergency care in a low-income country. Methods: Patients presenting to the emergency department with suspected pneumonia were evaluated with bedside lung ultrasound, single posterioranterior chest radiograph, and computed tomography (CT). Using CT as the gold standard, the sensitivity of lung ultrasound was compared to chest X-ray for the diagnosis of pneumonia using McNemar’s test for paired samples. Diagnostic characteristics for each test were calculated. Results: Of 62 patients included in the study, 44 (71%) were diagnosed with pneumonia by CT. Lung ultrasound demonstrated a sensitivity of 91% compared to chest X-ray which had a sensitivity of 73% (p = 0.01). Specificity of lung ultrasound and chest X-ray were 61 and 50% respectively. Conclusions: Bedside lung ultrasound demonstrated better sensitivity than chest X-ray for the diagnosis of pneumonia in Nepal. Trial registration: ClinicalTrials.gov, registration number NCT02949141. Registered 31 October 2016. -

ICD-10 Codes Are a Valid Tool for Identification of Pneumonia In
Epidemiol. Infect. (2008), 136, 232–240. f 2007 Cambridge University Press doi:10.1017/S0950268807008564 Printed in the United Kingdom ICD-10 codes are a valid tool for identification of pneumonia in hospitalized patients aged o65 years S. A. SKULL 1,2*, R. M. ANDREWS 2, G.B. BYRNES3, D.A. CAMPBELL3,4, 3 5 3,6 T. M. NOLAN , G.V. BROWN AND H.A. KELLY 1 Department of Paediatrics, University of Melbourne, Royal Children’s Hospital, Parkville, Victoria, Australia 2 Menzies School of Health Research, Darwin, Northern Territory, Australia 3 School of Population Health, University of Melbourne, Victoria, Australia 4 Monash Institute of Health Services Research, Monash Medical Centre, Clayton, Victoria, Australia 5 Department of Medicine, University of Melbourne, Royal Melbourne Hospital, Parkville, Victoria, Australia 6 Victorian Infectious Diseases Reference Laboratory, North Melbourne, Victoria, Australia (Accepted 28 March 2007; first published online 20 April 2007) SUMMARY This study examines the validity of using ICD-10 codes to identify hospitalized pneumonia cases. Using a case-cohort design, subjects were randomly selected from monthly cohorts of patients aged o65 years discharged from April 2000 to March 2002 from two large tertiary Australian hospitals. Cases had ICD-10-AM codes J10–J18 (pneumonia); the cohort sample was randomly selected from all discharges, frequency matched to cases by month. Codes were validated against three comparators: medical record notation of pneumonia, chest radiograph (CXR) report and both. Notation of pneumonia was determined for 5098/5101 eligible patients, and CXR reports reviewed for 3349/3464 (97%) patients with a CXR. Coding performed best against notation of pneumonia: kappa 0.95, sensitivity 97.8% (95% CI 97.1–98.3), specificity 96.9% (95% CI 96.2–97.5), positive predictive value (PPV) 96.2% (95% CI 95.4–97.0) and negative predictive value (NPV) 98.2% (95% CI 97.6–98.6). -

Assessment of High Resolution Computed Tomography in the Diagnosis of Interstitial Lung Disease
International Journal of Research in Medical Sciences Ayush M et al. Int J Res Med Sci. 2018 Jul;6(7):2251-2255 www.msjonline.org pISSN 2320-6071 | eISSN 2320-6012 DOI: http://dx.doi.org/10.18203/2320-6012.ijrms20182403 Original Research Article Assessment of high resolution computed tomography in the diagnosis of interstitial lung disease Mridul Ayush1*, Ishita Jakhanwal2 1Department of Radio-Diagnosis and Imaging, Dr. D. Y. Patil Medical College, Pimpri, Pune, Maharashtra, India 2Director, Ayucare Diagnostic Centre, Pune, Maharashtra, India Received: 15 May 2018 Accepted: 21 May 2018 *Correspondence: Dr. Mridul Ayush, E-mail: [email protected] Copyright: © the author(s), publisher and licensee Medip Academy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Background: Interstitial lung disease (ILD) are group of pulmonary disorders characterized by inflammation and fibrosis of gas exchanging portion of the lung and diffuse abnormalities on lung radiograph. Conventional computerized tomography plays a limited role in evaluation of interstitial lung disease due to its inability to demonstrate fine parenchymal details. High Resolution Computed Tomography (HRCT) is currently the most accurate non invasive modality for evaluating lung- parenchyma. So, the purpose of the study was to assess high resolution computed tomography in the diagnosis of interstitial lung disease. Methods: 50 patients with clinical suspicion of interstitial lung disease who were referred to Department of Radio- Diagnosis and Imaging for diagnosis and evaluation were subjected to both conventional radiography and HRCT. -

Role of Chest X-Ray with Ultrasonography in Evaluation of Pneumonia in an Icu Setting: a Prospective Study
ARC Journal of Radiology and Medical Imaging Volume 4, Issue 2, 2019, PP 1 -14 www.arcjournals.org Role of Chest X-Ray with Ultrasonography in Evaluation of Pneumonia in an Icu Setting: A Prospective Study Dr. Som Biswas* DMRD, M.S., Dr. Satish Pande. M.D., D.N.B, Ph.D. Department of Radiodiagnosis, King Edward Memorial Hospital, Pune, India *Corresponding Author: Som Biswas, Resident, Department of Radio diagnosis, KEM Pune, India Email: [email protected] Abstract Aim: Evaluation of role of Chest X-ray & Ultrasonography in pneumonia in an ICU setting. Introduction: Critically ill patients frequently need thoracic imaging due to dynamic nature of their clinical conditions. I studied the role of Chest X-ray & Ultrasonography in patient with pneumonia in an ICU setting in KEM Hospital, Pune. Materials and Methods: The present study was carried out at KEM Hospital, Pune in the Intensive Care Unit from 1st June 2018 to 31st May 2019 on patients with findings of pneumonia. Sample size: 150 patients as per calculation Summary and Conclusion Lung sonography was able to evaluate homogeneous opacities, air bronchogram, Kerley- A line, Kerley- B line, fluid in pleural space. The limitations of ultrasonography were non visualization of inhomogeneous opacities and volume loss. Thus lung sonography is complimentary for evaluation of pneumonia along with chest radiography. 1. INTRODUCTION cause of death worldwidede, preceded only by ischemic heart disease and cerebrovascular Critically ill patients frequently need thoracic diseases. It is the leading infectious cause of imaging due to dynamic nature of their clinical death and one of the most common reasons for conditions. -
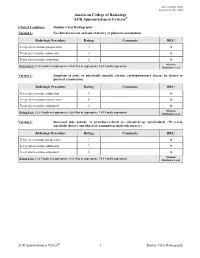
Routine Chest Radiography Variant 1: No Clinical Concern on Basis of History Or Physical Examination
Date of origin: 2000 Last review date: 2015 American College of Radiology ACR Appropriateness Criteria® Clinical Condition: Routine Chest Radiography Variant 1: No clinical concern on basis of history or physical examination. Radiologic Procedure Rating Comments RRL* X-ray chest routine preoperative 3 ☢ X-ray chest routine admission 3 ☢ X-ray chest routine outpatient 2 ☢ *Relative Rating Scale: 1,2,3 Usually not appropriate; 4,5,6 May be appropriate; 7,8,9 Usually appropriate Radiation Level Variant 2: Suspicion of acute or potentially unstable chronic cardiopulmonary disease by history or physical examination. Radiologic Procedure Rating Comments RRL* X-ray chest routine admission 9 ☢ X-ray chest routine preoperative 8 ☢ X-ray chest routine outpatient 8 ☢ *Relative Rating Scale: 1,2,3 Usually not appropriate; 4,5,6 May be appropriate; 7,8,9 Usually appropriate Radiation Level Variant 3: Increased risk, patient- or procedure-related (ie, advanced age [particularly >70 years], unreliable history and physical examination, high-risk surgery). Radiologic Procedure Rating Comments RRL* X-ray chest routine preoperative 7 ☢ X-ray chest routine admission 7 ☢ X-ray chest routine outpatient 6 ☢ *Relative Rating Scale: 1,2,3 Usually not appropriate; 4,5,6 May be appropriate; 7,8,9 Usually appropriate Radiation Level ACR Appropriateness Criteria® 1 Routine Chest Radiography ROUTINE CHEST RADIOGRAPHY Expert Panel on Thoracic Imaging: Barbara L. McComb, MD1; Jonathan H. Chung, MD2; Traves D. Crabtree, MD3; Darel E. Heitkamp, MD4; Mark D. Iannettoni, MD5; Clinton Jokerst, MD6; Anthony G. Saleh, MD7; Rakesh D. Shah, MD8; Robert M. Steiner, MD9; Tan-Lucien H. Mohammed, MD10; James G. -

The Role of High Resolution Computed Tomography in the Diagnosis of Interstitial Lung Disease
Thorax 1991;46:77-84 77 THORAX Thorax: first published as 10.1136/thx.46.2.77 on 1 February 1991. Downloaded from Review The role of high resolution computed tomography in the diagnosis of interstitial lung disease David M Hansell, Ian H Kerr Until recently the chest radiograph has been study. This protocol is widely used when a the only imaging technique used in the assess- comprehensive examination of the lungs is ment of patients with suspected diffuse lung required-for example, in the search for disease. In this context the chest radiograph is metastases. The volume averaging that occurs less than ideal. Problems arise with both false within the 1 cm thickness of the scan, negative and false positive results. It is well however, substantially reduces the ability of established that the chest radiograph may conventional computed tomography to resolve appear entirely normal in up to 10% of small structures. For high resolution com- patients with biopsy proved diffuse lung dis- puted tomography the section thickness is ease of various causes,1 and a poor quality reduced to 1-3 mm and a different software chest radiograph, especially of an obese reconstruction of the image is used to improve patient, may misleadingly raise the spectre of spatial resolution (figs la and lb). These scans diffuse lung disease. The two dimensional are interspaced by at least 1 cm. Where 3 mm nature of a chest radiograph dictates that there sections are taken 1 cm apart the radiation is superimposition of structures over the lungs dose to the breast is reduced to about half that and it has been estimated that up to 40% of of conventional computed tomography. -

Magnetic Resonance Imaging of Chest Wall Lesions
MAGNETIC RESONANCE IMAGING OF CHEST WALL LESIONS James D. Collins, MD, Marla Shaver, MD, Poonam Batra, MD, Kathleen Brown, MD, and Anthony C. Disher, MD Los Angeles, California Magnetic resonance imaging (MRI) demon- Clinical observations bring questions of whether strates surface anatomy, nerves, and soft models of radiological pathological correlation can be tissue pathology. Selective placement of the constructed to test the observations. 1-4 A thesis or cursor lines in MRI displays specific anatomy. theory is challenged by deriving a protocol. The The MR images can then be used as an adjunct radiologist who takes the opportunity to observe the in teaching surface anatomy to medical stu- clinical and pathological environment of the academic dents and to other health professionals. Be- arena will enhance teaching and research within the cause the normal surface anatomy could be clinical setting. Observations may come from reading imaged at UCLA's radiology department, it was an endless number of radiographs or surgical operative decided to image soft tissue abnormalities with reports, or from performing special procedures.3'5'6 MR to assist in patient care. A series of tests may be designed for animal Patients imaged were scheduled for special research,37 or data may be stored based on patient procedures of the chest or staging lymphangi- observation. The information is recorded, and statistics ograms. Patients were placed into categories are reviewed to evaluate the theory. If the theory is valid depending on known diagnosis or interesting and can be modified for patient care, the procedure may clinical presentation. The diagnostic catego- be adopted.7-9 ries included Hodgkin's disease, melanoma, Magnetic resonance imaging (MRI) demonstrates carcinomas (eg, lung or breast), lymphedema, surface anatomy, nerves, and soft tissue pathology.10-13 sarcomas, dermatological disorders, and neu- Selective placement of cursor lines in MRI displays rological disorders. -

Acr–Spr–Str Practice Parameter for the Performance of Portable (Mobile Unit) Chest Radiography
The American College of Radiology, with more than 30,000 members, is the principal organization of radiologists, radiation oncologists, and clinical medical physicists in the United States. The College is a nonprofit professional society whose primary purposes are to advance the science of radiology, improve radiologic services to the patient, study the socioeconomic aspects of the practice of radiology, and encourage continuing education for radiologists, radiation oncologists, medical physicists, and persons practicing in allied professional fields. The American College of Radiology will periodically define new practice parameters and technical standards for radiologic practice to help advance the science of radiology and to improve the quality of service to patients throughout the United States. Existing practice parameters and technical standards will be reviewed for revision or renewal, as appropriate, on their fifth anniversary or sooner, if indicated. Each practice parameter and technical standard, representing a policy statement by the College, has undergone a thorough consensus process in which it has been subjected to extensive review and approval. The practice parameters and technical standards recognize that the safe and effective use of diagnostic and therapeutic radiology requires specific training, skills, and techniques, as described in each document. Reproduction or modification of the published practice parameter and technical standard by those entities not providing these services is not authorized. Revised 2017 (Resolution 3)* ACR–SPR–STR PRACTICE PARAMETER FOR THE PERFORMANCE OF PORTABLE (MOBILE UNIT) CHEST RADIOGRAPHY PREAMBLE This document is an educational tool designed to assist practitioners in providing appropriate radiologic care for patients. Practice Parameters and Technical Standards are not inflexible rules or requirements of practice and are not intended, nor should they be used, to establish a legal standard of care1. -

Near-Drowning
Central Journal of Trauma and Care Bringing Excellence in Open Access Review Article *Corresponding author Bhagya Sannananja, Department of Radiology, University of Washington, 1959, NE Pacific St, Seattle, WA Near-Drowning: Epidemiology, 98195, USA, Tel: 830-499-1446; Email: Submitted: 23 May 2017 Pathophysiology and Imaging Accepted: 19 June 2017 Published: 22 June 2017 Copyright Findings © 2017 Sannananja et al. Carlos S. Restrepo1, Carolina Ortiz2, Achint K. Singh1, and ISSN: 2573-1246 3 Bhagya Sannananja * OPEN ACCESS 1Department of Radiology, University of Texas Health Science Center at San Antonio, USA Keywords 2Department of Internal Medicine, University of Texas Health Science Center at San • Near-drowning Antonio, USA • Immersion 3Department of Radiology, University of Washington, USA\ • Imaging findings • Lung/radiography • Magnetic resonance imaging Abstract • Tomography Although occasionally preventable, drowning is a major cause of accidental death • X-Ray computed worldwide, with the highest rates among children. A new definition by WHO classifies • Nonfatal drowning drowning as the process of experiencing respiratory impairment from submersion/ immersion in liquid, which can lead to fatal or nonfatal drowning. Hypoxemia seems to be the most severe pathophysiologic consequence of nonfatal drowning. Victims may sustain severe organ damage, mainly to the brain. It is difficult to predict an accurate neurological prognosis from the initial clinical presentation, laboratory and radiological examinations. Imaging plays an important role in the diagnosis and management of near-drowning victims. Chest radiograph is commonly obtained as the first imaging modality, which usually shows perihilar bilateral pulmonary opacities; yet 20% to 30% of near-drowning patients may have normal initial chest radiographs. Brain hypoxia manifest on CT by diffuse loss of gray-white matter differentiation, and on MRI diffusion weighted sequence with high signal in the injured regions. -

Magnetic Resonance Imaging As an Adjunct to Computed Tomography In
Published online: 2021-07-26 THORACIC IMAGING Magnetic resonance imaging as an adjunct to computed tomography in the diagnosis of pulmonary Hydatid cysts Roopa Tandur, Aparna Irodi, Binita Riya Chacko, Leena Robinson Vimala, Devasahayam Jesudas Christopher1, Birla Roy Gnanamuthu2 Departments of Radiodiagnosis, 1Pulmonary Medicine and 2Cardiothoracic Surgery, Christian Medical College, Vellore, Tamil Nadu, India Correspondence: Dr Aparna Irodi, Department of Radiodiagnosis, Christian Medical College, Vellore, Tamil Nadu, India. E‑mail: [email protected] Abstract Introduction: Although pulmonary hydatid cysts can be diagnosed on computed tomography (CT), sometimes findings can be atypical. Other hypodense infective or neoplastic lesions may mimic hydatid cysts. We proposed that magnetic resonance imaging (MRI) may act as a problem‑solving tool, aiding the definite diagnosis of hydatid cysts and differentiating it from its mimics. The aim of this study is to assess the findings of pulmonary hydatid cysts on CT and MRI and the additional contribution of MRI in doubtful cases. Materials and Methods: This is a retrospective study of 90 patients with suspected hydatid cysts. CT and MRI findings were noted and role of MRI in diagnosing hydatid cysts and its mimics was studied. Descriptive statistics for CT findings and sensitivity and specificity of MRI were calculated using surgery or histopathology as gold standard.Results: Of the 90 patients with suspected pulmonary hydatid cysts, there were 52 true‑positive and 7 false‑positive cases on CT. Commonest CT finding was unilocular thick‑walled cyst. In the 26 patients who had additional MRI, based on T2‑weighted hypointense rim or folded membranes, accurate preoperative differentiation of 14 patients with hydatid cysts from 10 patients with alternate diagnosis was possible.