The Fragmentation of Gondwanaland: Influence on the Historical Biogeography and Morphological Evolution Within Dragon Lizards (Squamata: Agamidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogenetic Relationships and Subgeneric Taxonomy of Toad�Headed Agamas Phrynocephalus (Reptilia, Squamata, Agamidae) As Determined by Mitochondrial DNA Sequencing E
ISSN 00124966, Doklady Biological Sciences, 2014, Vol. 455, pp. 119–124. © Pleiades Publishing, Ltd., 2014. Original Russian Text © E.N. Solovyeva, N.A. Poyarkov, E.A. Dunayev, R.A. Nazarov, V.S. Lebedev, A.A. Bannikova, 2014, published in Doklady Akademii Nauk, 2014, Vol. 455, No. 4, pp. 484–489. GENERAL BIOLOGY Phylogenetic Relationships and Subgeneric Taxonomy of ToadHeaded Agamas Phrynocephalus (Reptilia, Squamata, Agamidae) as Determined by Mitochondrial DNA Sequencing E. N. Solovyeva, N. A. Poyarkov, E. A. Dunayev, R. A. Nazarov, V. S. Lebedev, and A. A. Bannikova Presented by Academician Yu.Yu. Dgebuadze October 25, 2013 Received October 30, 2013 DOI: 10.1134/S0012496614020148 Toadheaded agamas (Phrynocephalus) is an essen Trapelus, and Stellagama) were used in molecular tial element of arid biotopes throughout the vast area genetic analysis. In total, 69 sequences from the Gen spanning the countries of Middle East and Central Bank were studied, 28 of which served as outgroups (the Asia. They constitute one of the most diverse genera of members of Agamidae, Chamaeleonidae, Iguanidae, the agama family (Agamidae), variously estimated to and Lacertidae). comprise 26 to 40 species [1]. The subgeneric Phryno The fragment sequences of the following four cephalus taxonomy is poorly studied: recent taxo mitochondrial DNA genes were used in phylogenetic nomic revision have been conducted without analysis analysis: the genes of subunit I of cytochrome c oxi of the entire genus diversity [1]; therefore, its phyloge dase (COI), of subunits II and IV of NADHdehydro netic position within Agamidae family remains genase (ND2 and ND4), and of cytochrome b (cyt b). -

Gliding Dragons and Flying Squirrels: Diversifying Versus Stabilizing Selection on Morphology Following the Evolution of an Innovation
vol. 195, no. 2 the american naturalist february 2020 E-Article Gliding Dragons and Flying Squirrels: Diversifying versus Stabilizing Selection on Morphology following the Evolution of an Innovation Terry J. Ord,1,* Joan Garcia-Porta,1,† Marina Querejeta,2,‡ and David C. Collar3 1. Evolution and Ecology Research Centre and the School of Biological, Earth and Environmental Sciences, University of New South Wales, Kensington, New South Wales 2052, Australia; 2. Institute of Evolutionary Biology (CSIC–Universitat Pompeu Fabra), Passeig Marítim de la Barceloneta, 37–49, Barcelona 08003, Spain; 3. Department of Organismal and Environmental Biology, Christopher Newport University, Newport News, Virginia 23606 Submitted August 1, 2018; Accepted July 16, 2019; Electronically published December 17, 2019 Online enhancements: supplemental material. Dryad data: https://doi.org/10.5061/dryad.t7g227h. fi abstract: Evolutionary innovations and ecological competition are eral de nitions of what represents an innovation have been factors often cited as drivers of adaptive diversification. Yet many offered (reviewed by Rabosky 2017), this classical descrip- innovations result in stabilizing rather than diversifying selection on tion arguably remains the most useful (Galis 2001; Stroud morphology, and morphological disparity among coexisting species and Losos 2016; Rabosky 2017). Hypothesized innovations can reflect competitive exclusion (species sorting) rather than sympat- have drawn considerable attention among ecologists and ric adaptive divergence (character displacement). We studied the in- evolutionary biologists because they can expand the range novation of gliding in dragons (Agamidae) and squirrels (Sciuridae) of ecological niches occupied within communities. In do- and its effect on subsequent body size diversification. We found that gliding either had no impact (squirrels) or resulted in strong stabilizing ing so, innovations are thought to be important engines of selection on body size (dragons). -

Fossil Amphibians and Reptiles from the Neogene Locality of Maramena (Greece), the Most Diverse European Herpetofauna at the Miocene/Pliocene Transition Boundary
Palaeontologia Electronica palaeo-electronica.org Fossil amphibians and reptiles from the Neogene locality of Maramena (Greece), the most diverse European herpetofauna at the Miocene/Pliocene transition boundary Georgios L. Georgalis, Andrea Villa, Martin Ivanov, Davit Vasilyan, and Massimo Delfino ABSTRACT We herein describe the fossil amphibians and reptiles from the Neogene (latest Miocene or earliest Pliocene; MN 13/14) locality of Maramena, in northern Greece. The herpetofauna is shown to be extremely diverse, comprising at least 30 different taxa. Amphibians include at least six urodelan (Cryptobranchidae indet., Salamandrina sp., Lissotriton sp. [Lissotriton vulgaris group], Lissotriton sp., Ommatotriton sp., and Sala- mandra sp.), and three anuran taxa (Latonia sp., Hyla sp., and Pelophylax sp.). Rep- tiles are much more speciose, being represented by two turtle (the geoemydid Mauremys aristotelica and a probable indeterminate testudinid), at least nine lizard (Agaminae indet., Lacertidae indet., ?Lacertidae indet., aff. Palaeocordylus sp., ?Scin- cidae indet., Anguis sp., five morphotypes of Ophisaurus, Pseudopus sp., and at least one species of Varanus), and 10 snake taxa (Scolecophidia indet., Periergophis micros gen. et sp. nov., Paraxenophis spanios gen. et sp. nov., Hierophis cf. hungaricus, another distinct “colubrine” morphotype, Natrix aff. rudabanyaensis, and another dis- tinct species of Natrix, Naja sp., cf. Micrurus sp., and a member of the “Oriental Vipers” complex). The autapomorphic features and bizarre vertebral morphology of Perier- gophis micros gen. et sp. nov. and Paraxenophis spanios gen. et sp. nov. render them readily distinguishable among fossil and extant snakes. Cryptobranchids, several of the amphibian genera, scincids, Anguis, Pseudopus, and Micrurus represent totally new fossil occurrences, not only for the Greek area, but for the whole southeastern Europe. -

A New Subspecies of Jackyhosersaur Hoser, 2013 from North-West Australia
Australasian Journal of Herpetology 5 Australasian Journal of Herpetology 49:5-6. Published 6 August 2020. ISSN 1836-5698 (Print) ISSN 1836-5779 (Online) A new subspecies of Jackyhosersaur Hoser, 2013 from north-west Australia. LSIDURN:LSID:ZOOBANK.ORG:PUB:2877185D-E93C-4040-973C-89657DC512B1 RAYMOND T. HOSER LSID urn:lsid:zoobank.org:author:F9D74EB5-CFB5-49A0-8C7C-9F993B8504AE 488 Park Road, Park Orchards, Victoria, 3134, Australia. Phone: +61 3 9812 3322 Fax: 9812 3355 E-mail: snakeman (at) snakeman.com.au Received 10 February 2017, Accepted 17 January 2020, Published 6 August 2020. ABSTRACT As part of an ongoing audit of Australian reptiles, specimens of the endemic Western Australian dragon species, Jackyhosersaur superba (Storr, 1974), known to most herpetologists as “Diporiphora superba Storr, 1974” were inspected from all parts of the known range. A population from the Manning Creek Gorge area of the Kimberley in Western Australia were found to be morphologically divergent and so are formally described herein as a new subspecies. Keywords: Lizards; taxonomy; nomenclature; Agamidae; Western Australia; Diporiphora; Jackyhosersaur; superba; newsubspecies; jackyhoserae. INTRODUCTION Jackyhosersaur superba (Storr, 1974) as currently recognized, As stated in the abstract, as part of an ongoing audit of even though all occur within close geographical proximity, being Australian reptiles, specimens of the endemic Western restricted to the high rainfall parts of the West Kimberley Region. Australian dragon species, Jackyhosersaur superba (Storr, Significant is that the species appears to be reasonably 1974) were inspected from all parts of the known range. A common and different populations are in close proximity to one population from the Manning Creek Gorge area of the Kimberley another without obvious biogeographical barriers stopping gene in Western Australia were found to be morphologically divergent flow between them. -

Herpetological Review
Herpetological Review Volume 41, Number 2 — June 2010 SSAR Offi cers (2010) HERPETOLOGICAL REVIEW President The Quarterly News-Journal of the Society for the Study of Amphibians and Reptiles BRIAN CROTHER Department of Biological Sciences Editor Southeastern Louisiana University ROBERT W. HANSEN Hammond, Louisiana 70402, USA 16333 Deer Path Lane e-mail: [email protected] Clovis, California 93619-9735, USA [email protected] President-elect JOSEPH MENDLELSON, III Zoo Atlanta, 800 Cherokee Avenue, SE Associate Editors Atlanta, Georgia 30315, USA e-mail: [email protected] ROBERT E. ESPINOZA KERRY GRIFFIS-KYLE DEANNA H. OLSON California State University, Northridge Texas Tech University USDA Forestry Science Lab Secretary MARION R. PREEST ROBERT N. REED MICHAEL S. GRACE PETER V. LINDEMAN USGS Fort Collins Science Center Florida Institute of Technology Edinboro University Joint Science Department The Claremont Colleges EMILY N. TAYLOR GUNTHER KÖHLER JESSE L. BRUNNER Claremont, California 91711, USA California Polytechnic State University Forschungsinstitut und State University of New York at e-mail: [email protected] Naturmuseum Senckenberg Syracuse MICHAEL F. BENARD Treasurer Case Western Reserve University KIRSTEN E. NICHOLSON Department of Biology, Brooks 217 Section Editors Central Michigan University Mt. Pleasant, Michigan 48859, USA Book Reviews Current Research Current Research e-mail: [email protected] AARON M. BAUER JOSHUA M. HALE BEN LOWE Department of Biology Department of Sciences Department of EEB Publications Secretary Villanova University MuseumVictoria, GPO Box 666 University of Minnesota BRECK BARTHOLOMEW Villanova, Pennsylvania 19085, USA Melbourne, Victoria 3001, Australia St Paul, Minnesota 55108, USA P.O. Box 58517 [email protected] [email protected] [email protected] Salt Lake City, Utah 84158, USA e-mail: [email protected] Geographic Distribution Geographic Distribution Geographic Distribution Immediate Past President ALAN M. -

NHBSS 061 1G Hikida Fieldg
Book Review N$7+IST. BULL. S,$0 SOC. 61(1): 41–51, 2015 A Field Guide to the Reptiles of Thailand by Tanya Chan-ard, John W. K. Parr and Jarujin Nabhitabhata. Oxford University Press, New York, 2015. 344 pp. paper. ISBN: 9780199736492. 7KDLUHSWLOHVZHUHÀUVWH[WHQVLYHO\VWXGLHGE\WZRJUHDWKHUSHWRORJLVWV0DOFROP$UWKXU 6PLWKDQG(GZDUG+DUULVRQ7D\ORU7KHLUFRQWULEXWLRQVZHUHSXEOLVKHGDV6MITH (1931, 1935, 1943) and TAYLOR 5HFHQWO\RWKHUERRNVDERXWUHSWLOHVDQGDPSKLELDQV LQ7KDLODQGZHUHSXEOLVKHG HJ&HAN-ARD ET AL., 1999: COX ET AL DVZHOODVPDQ\ SDSHUV+RZHYHUWKHVHERRNVZHUHWD[RQRPLFVWXGLHVDQGQRWJXLGHVIRURUGLQDU\SHRSOH7ZR DGGLWLRQDOÀHOGJXLGHERRNVRQUHSWLOHVRUDPSKLELDQVDQGUHSWLOHVKDYHDOVREHHQSXEOLVKHG 0ANTHEY & GROSSMANN, 1997; DAS EXWWKHVHERRNVFRYHURQO\DSDUWRIWKHIDXQD The book under review is very well prepared and will help us know Thai reptiles better. 2QHRIWKHDXWKRUV-DUXMLQ1DEKLWDEKDWDZDVP\ROGIULHQGIRUPHUO\WKH'LUHFWRURI1DWXUDO +LVWRU\0XVHXPWKH1DWLRQDO6FLHQFH0XVHXP7KDLODQG+HZDVDQH[FHOOHQWQDWXUDOLVW DQGKDGH[WHQVLYHNQRZOHGJHDERXW7KDLDQLPDOVHVSHFLDOO\DPSKLELDQVDQGUHSWLOHV,Q ZHYLVLWHG.KDR6RL'DR:LOGOLIH6DQFWXDU\WRVXUYH\KHUSHWRIDXQD+HDGYLVHGXV WRGLJTXLFNO\DURXQGWKHUH:HFROOHFWHGIRXUVSHFLPHQVRIDibamusZKLFKZHGHVFULEHG DVDQHZVSHFLHVDibamus somsaki +ONDA ET AL 1RZ,DPYHU\JODGWRNQRZWKDW WKLVERRNZDVSXEOLVKHGE\KLPDQGKLVFROOHDJXHV8QIRUWXQDWHO\KHSDVVHGDZD\LQ +LVXQWLPHO\GHDWKPD\KDYHGHOD\HGWKHSXEOLFDWLRQRIWKLVERRN7KHERRNLQFOXGHVQHDUO\ DOOQDWLYHUHSWLOHV PRUHWKDQVSHFLHV LQ7KDLODQGDQGPRVWSLFWXUHVZHUHGUDZQZLWK H[FHOOHQWGHWDLO,WLVDYHU\JRRGÀHOGJXLGHIRULGHQWLÀFDWLRQRI7KDLUHSWLOHVIRUVWXGHQWV -

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

Download Download
HAMADRYAD Vol. 27. No. 2. August, 2003 Date of issue: 31 August, 2003 ISSN 0972-205X CONTENTS T. -M. LEONG,L.L.GRISMER &MUMPUNI. Preliminary checklists of the herpetofauna of the Anambas and Natuna Islands (South China Sea) ..................................................165–174 T.-M. LEONG & C-F. LIM. The tadpole of Rana miopus Boulenger, 1918 from Peninsular Malaysia ...............175–178 N. D. RATHNAYAKE,N.D.HERATH,K.K.HEWAMATHES &S.JAYALATH. The thermal behaviour, diurnal activity pattern and body temperature of Varanus salvator in central Sri Lanka .........................179–184 B. TRIPATHY,B.PANDAV &R.C.PANIGRAHY. Hatching success and orientation in Lepidochelys olivacea (Eschscholtz, 1829) at Rushikulya Rookery, Orissa, India ......................................185–192 L. QUYET &T.ZIEGLER. First record of the Chinese crocodile lizard from outside of China: report on a population of Shinisaurus crocodilurus Ahl, 1930 from north-eastern Vietnam ..................193–199 O. S. G. PAUWELS,V.MAMONEKENE,P.DUMONT,W.R.BRANCH,M.BURGER &S.LAVOUÉ. Diet records for Crocodylus cataphractus (Reptilia: Crocodylidae) at Lake Divangui, Ogooué-Maritime Province, south-western Gabon......................................................200–204 A. M. BAUER. On the status of the name Oligodon taeniolatus (Jerdon, 1853) and its long-ignored senior synonym and secondary homonym, Oligodon taeniolatus (Daudin, 1803) ........................205–213 W. P. MCCORD,O.S.G.PAUWELS,R.BOUR,F.CHÉROT,J.IVERSON,P.C.H.PRITCHARD,K.THIRAKHUPT, W. KITIMASAK &T.BUNDHITWONGRUT. Chitra burmanica sensu Jaruthanin, 2002 (Testudines: Trionychidae): an unavailable name ............................................................214–216 V. GIRI,A.M.BAUER &N.CHATURVEDI. Notes on the distribution, natural history and variation of Hemidactylus giganteus Stoliczka, 1871 ................................................217–221 V. WALLACH. -

Zoology ABSTRACT
Research Paper Volume : 3 | Issue : 9 | September 2014 • ISSN No 2277 - 8179 New record of Roux’s Forest Lizard Calotes Zoology rouxii (Duméril & Bibron, 1837) (Reptilia: KEYWORDS : Calotes rouxii, range exten- Squamata: Agamidae) from Sandur and sion, distribution update, Karnataka Gulbarga, Karnataka, India with a note on its known distribution Biodiversity Research and Conservation Society, 303 Orchid, Sri Sai Nagar Colony, Aditya Srinivasulu Kanajiguda, Secunderabad, Telangana 500 015, India. Natural History Museum and Wildlife Biology and Taxonomy Lab, Department of Zoology, * C. Srinivasulu University College of Science, Osmania University, Hyderabad, Telangana 500 007, India. *Corresponding Author ABSTRACT Roux’s Forest Lizard Calotes rouxii (Duméril & Bibron, 1837) is chiefly a forest-dwelling draconine agamid that is widely distributed in the Western Ghats and occasionally reported from the Eastern Ghats and other localities in the central peninsular India. We report the presence of this species for the first time from Sandur forests in Bellary district, Karnataka based on a voucher specimen and from Gulbarga township based on sighting record. A detailed distribution map showing localities from where the species is known is also provided. The genus Calotes Daudin, 1802, belonging to the draconian fam- ized by the presence of a dewlap in males, two slender spines on ily Agamidae (Reptilia) is native to South Asia, South-East Asia either side of the head and a dark groove before the shoulder. and Southern China. It is represented -

F3999f15-C572-46Ad-Bbbe
THE STATUTES OF THE REPUBLIC OF SINGAPORE ENDANGERED SPECIES (IMPORT AND EXPORT) ACT (CHAPTER 92A) (Original Enactment: Act 5 of 2006) REVISED EDITION 2008 (1st January 2008) Prepared and Published by THE LAW REVISION COMMISSION UNDER THE AUTHORITY OF THE REVISED EDITION OF THE LAWS ACT (CHAPTER 275) Informal Consolidation – version in force from 22/6/2021 CHAPTER 92A 2008 Ed. Endangered Species (Import and Export) Act ARRANGEMENT OF SECTIONS PART I PRELIMINARY Section 1. Short title 2. Interpretation 3. Appointment of Director-General and authorised officers PART II CONTROL OF IMPORT, EXPORT, ETC., OF SCHEDULED SPECIES 4. Restriction on import, export, etc., of scheduled species 5. Control of scheduled species in transit 6. Defence to offence under section 4 or 5 7. Issue of permit 8. Cancellation of permit PART III ENFORCEMENT POWERS AND PROCEEDINGS 9. Power of inspection 10. Power to investigate and require information 11. Power of entry, search and seizure 12. Powers ancillary to inspections and searches 13. Power to require scheduled species to be marked, etc. 14. Power of arrest 15. Forfeiture 16. Obstruction 17. Penalty for false declarations, etc. 18. General penalty 19. Abetment of offences 20. Offences by bodies corporate, etc. 1 Informal Consolidation – version in force from 22/6/2021 Endangered Species (Import and 2008 Ed. Export) CAP. 92A 2 PART IV MISCELLANEOUS Section 21. Advisory Committee 22. Fees, etc., payable to Board 23. Board not liable for damage caused to goods or property as result of search, etc. 24. Jurisdiction of court, etc. 25. Composition of offences 26. Exemption 27. Service of documents 28. -
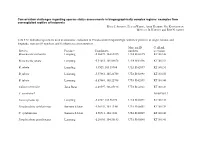
Conservation Challenges Regarding Species Status Assessments in Biogeographically Complex Regions: Examples from Overexploited Reptiles of Indonesia KYLE J
Conservation challenges regarding species status assessments in biogeographically complex regions: examples from overexploited reptiles of Indonesia KYLE J. SHANEY, ELIJAH WOSTL, AMIR HAMIDY, NIA KURNIAWAN MICHAEL B. HARVEY and ERIC N. SMITH TABLE S1 Individual specimens used in taxonomic evaluation of Pseudocalotes tympanistriga, with their province of origin, latitude and longitude, museum ID numbers, and GenBank accession numbers. Museum ID GenBank Species Province Coordinates numbers accession Bronchocela cristatella Lampung -5.36079, 104.63215 UTA R 62895 KT180148 Bronchocela jubata Lampung -5.54653, 105.04678 UTA R 62896 KT180152 B. jubata Lampung -5.5525, 105.18384 UTA R 62897 KT180151 B. jubata Lampung -5.57861, 105.22708 UTA R 62898 KT180150 B. jubata Lampung -5.57861, 105.22708 UTA R 62899 KT180146 Calotes versicolor Jawa Barat -6.49597, 106.85198 UTA R 62861 KT180149 C. versicolor* NC009683.1 Gonocephalus sp. Lampung -5.2787, 104.56198 UTA R 60571 KT180144 Pseudocalotes cybelidermus Sumatra Selatan -4.90149, 104.13401 UTA R 60551 KT180139 P. cybelidermus Sumatra Selatan -4.90711, 104.1348 UTA R 60549 KT180140 Pseudocalotes guttalineatus Lampung -5.28105, 104.56183 UTA R 60540 KT180141 P. guttalineatus Sumatra Selatan -4.90681, 104.13457 UTA R 60501 KT180142 Pseudocalotes rhammanotus Lampung -4.9394, 103.85292 MZB 10804 KT180147 Pseudocalotes species 4 Sumatra Barat -2.04294, 101.31129 MZB 13295 KT211019 Pseudocalotes tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60544 KT180143 P. tympanistriga Jawa Barat -6.74181, 107.0061 UTA R 60547 KT180145 Pogona vitticeps* AB166795.1 *Entry to GenBank by previous authors TABLE S2 Reptile species currently believed to occur Java and Sumatra, Indonesia, with IUCN Red List status, and certainty of occurrence. -

COMMISSION REGULATION (EC) No 834/2004
L 127/40EN Official Journal of the European Union 29.4.2004 COMMISSION REGULATION (EC) No 834/2004 of 28 April 2004 amending Council Regulation (EC) No 338/97 on the protection of species of wild fauna and flora by regulating trade therein THE COMMISSION OF THE EUROPEAN COMMUNITIES, definition of ‘specimens’ given by Article 2(t) of Regu- lation (EC) No 338/97; the annotation regarding Aloe Having regard to the Treaty establishing the European spp. needs to make an explicit reference to the species Community, listed in Annex A; and the annotation to Guaiacum spp. Having regard to Council Regulation (EC) No 338/97 of 9 needs to be changed in order to designate the parts and December 1996 on the protection of species of wild fauna and derivatives decided upon at the 12th Conference. flora by regulating trade therein (1), and in particular Article 19(3) thereof, (5) The Scientific Review Group has established, on the basis of the criteria set out in Article 3(4)(a) of Regu- Whereas: lation (EC) No 338/97, that certain species must be with- drawn from the list of animals whose importation into (1) Council Regulation (EC) No 338/97 lists animal and the Community should, on account of the volume plant species in respect of which trade is restricted or involved, be monitored, whilst certain other species controlled. Those lists incorporate the lists set out in the must be added to that list. annexes to the Convention on International Trade in Endangered Species of Wild Fauna and Flora, hereinafter (6) Regulation (EC) No 338/97 should therefore be ‘the CITES Convention’.