Two Biological Constants for Accurate Classification and Evolution Pattern Analysis of Subgen.Strobus and Subgen
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
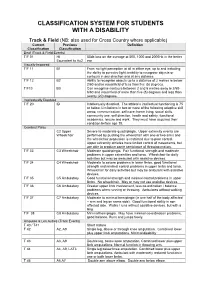
Disability Classification System
CLASSIFICATION SYSTEM FOR STUDENTS WITH A DISABILITY Track & Field (NB: also used for Cross Country where applicable) Current Previous Definition Classification Classification Deaf (Track & Field Events) T/F 01 HI 55db loss on the average at 500, 1000 and 2000Hz in the better Equivalent to Au2 ear Visually Impaired T/F 11 B1 From no light perception at all in either eye, up to and including the ability to perceive light; inability to recognise objects or contours in any direction and at any distance. T/F 12 B2 Ability to recognise objects up to a distance of 2 metres ie below 2/60 and/or visual field of less than five (5) degrees. T/F13 B3 Can recognise contours between 2 and 6 metres away ie 2/60- 6/60 and visual field of more than five (5) degrees and less than twenty (20) degrees. Intellectually Disabled T/F 20 ID Intellectually disabled. The athlete’s intellectual functioning is 75 or below. Limitations in two or more of the following adaptive skill areas; communication, self-care; home living, social skills, community use, self direction, health and safety, functional academics, leisure and work. They must have acquired their condition before age 18. Cerebral Palsy C2 Upper Severe to moderate quadriplegia. Upper extremity events are Wheelchair performed by pushing the wheelchair with one or two arms and the wheelchair propulsion is restricted due to poor control. Upper extremity athletes have limited control of movements, but are able to produce some semblance of throwing motion. T/F 33 C3 Wheelchair Moderate quadriplegia. Fair functional strength and moderate problems in upper extremities and torso. -

ASA East Region Para-Swimming Meet 2017 Condidtions 19.03.17
ASA EAST REGION PARA-SWIMMING MEET 2017 Level 3 Licence No. 3ER170675 Sunday 19th March 2017 Newmarket Leisure Centre, Exning Way, Newmarket, Suffolk, CB8 0EA Warm up: 5.00pm Start 5.30pm Meet Conditions 1. The gala is a Level 3 licensed meet and will be held under ASA Laws and Regulations, ASA Technical Rules of Racing and IPC Swimming Rules. 2. The Promoter is Jackie Harvey on behalf of the ASA East Region Disability Committee. Email all queries to [email protected] 3. The pool length is 25 metres, 6 lanes with Swiss electronic timing. 4. The competition is open to any swimmer with a disability: physical, sensory or learning impairments. 5. Each competitor shall be an ASA Category 2 member or an equivalent member of SASA or WASA whose membership record shows a S1-15, SB1–SB15 or SM1-SM15 IPC Swimming classification, which is held on the British Swimming or IPC Swimming Classification database at the time of entry. 6. Swimmers on the functional classification waiting list, or have an active application with UKSA or INAS, made within the last 12 months, are invited to compete. They must still meet the ASA Category 2 membership requirement. They would achieve a time in a licensed event but will not be eligible to receive medals. 7. Competitors should indicate on their entry their relevant: • IPC classification or British Swimming classification • UKSA or INAS classification (accompanied by the relevant number) • British Blind Sport classification • UK Deaf Sport classification • Exception codes Where swimmers are on the classification waiting list please insert W/L 8. -

Tokyo 2020 Paralympic Games
TOKYO 2020 PARALYMPIC GAMES QUALIFICATION REGULATIONS REVISED EDITION, APRIL 2021 INTERNATIONAL PARALYMPIC COMMITTEE 2 CONTENTS 1. Introduction 2. Tokyo 2020 Paralympic Games Programme Overview 3. General IPC Regulations on Eligibility 4. IPC Redistribution Policy of Vacant Qualification Slots 5. Universality Wild Cards 6. Key Dates 7. Archery 8. Athletics 9. Badminton 10. Boccia 11. Canoe 12. Cycling (Track and Road) 13. Equestrian 14. Football 5-a-side 15. Goalball 16. Judo 17. Powerlifting 18. Rowing 19. Shooting 20. Swimming 21. Table Tennis 22. Taekwondo 23. Triathlon 24. Volleyball (Sitting) 25. Wheelchair Basketball 26. Wheelchair Fencing 27. Wheelchair Rugby 28. Wheelchair Tennis 29. Glossary 30. Register of Updates INTERNATIONAL PARALYMPIC COMMITTEE 3 INTRODUCTION These Qualification Regulations (Regulations) describe in detail how athletes and teams can qualify for the Tokyo 2020 Paralympic Games in each of the twenty- two (22) sports on the Tokyo 2020 Paralympic Games Programme (Games Programme). It provides to the National Paralympic Committees (NPCs), to National Federations (NFs), to sports administrators, coaches and to the athletes themselves the conditions that allow participation in the signature event of the Paralympic Movement. These Regulations present: • an overview of the Games Programme; • the general IPC regulations on eligibility; • the specific qualification criteria for each sport (in alphabetical order); and • a glossary of the terminology used throughout the Regulations. STRUCTURE OF SPORT-SPECIFIC QUALIFICATION -

The Use of Next Generation Sequencing to Study the Environmental Mycobiome and Its Potential Health Effects
The use of next generation sequencing to study the environmental mycobiome and its potential health effects Emma Marczylo Bioaerosols – June 2017 Overview • Brief Background: • Why are CRCE interested in fungal bioaerosols? • Mycobiome analysis: • Why use next generation sequencing? • Ongoing work • What is the current focus of CRCE’s bioaerosol research? 2 Bioaerosols – June 2017 Why bioaerosols? • Respiratory health/toxicology a big focus within our department & bioaerosols represent a current respiratory health concern 3 Bioaerosols – June 2017 Why bioaerosols? • Common sources relevant to public health 4 Bioaerosols – June 2017 Why bioaerosols? • Public concern over health effects of living near composting and intensive farming sites • Systematic reviews on exposures and health outcomes related to bioaerosol emissions from composting facilities (published 2015*) or intensive farming (ongoing) in collaboration with SAHSU • Evidence on both exposure assessment and health effects are limited • A big unknown is the microbial composition of such bioaerosols *Pearson et al, 2015, J Toxicol Environ Health B Crit Rev,18:43-69 5 Bioaerosols – June 2017 Why fungi? Normally die rapidly due to water evaporation, although increased humidity and clumping can prolong survival • Can remain viable for much longer periods, even at low humidity & high/low temperatures • Much less known about the fungal composition of bioaerosols (and other samples) • Fungi linked with development and exacerbation of asthma symptoms 6 Bioaerosols – June 2017 Mycobiome -

Framework for In-Field Analyses of Performance and Sub-Technique Selection in Standing Para Cross-Country Skiers
sensors Article Framework for In-Field Analyses of Performance and Sub-Technique Selection in Standing Para Cross-Country Skiers Camilla H. Carlsen 1,*, Julia Kathrin Baumgart 1, Jan Kocbach 1,2, Pål Haugnes 1 , Evy M. B. Paulussen 1,3 and Øyvind Sandbakk 1 1 Centre for Elite Sports Research, Department of Neuromedicine and Movement Science, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, 7491 Trondheim, Norway; [email protected] (J.K.B.); [email protected] (J.K.); [email protected] (P.H.); [email protected] (E.M.B.P.); [email protected] (Ø.S.) 2 NORCE Norwegian Research Centre AS, 5008 Bergen, Norway 3 Faculty of Health, Medicine & Life Sciences, Maastricht University, 6200 MD Maastricht, The Netherlands * Correspondence: [email protected]; Tel.: +47-452-40-788 Abstract: Our aims were to evaluate the feasibility of a framework based on micro-sensor technology for in-field analyses of performance and sub-technique selection in Para cross-country (XC) skiing by using it to compare these parameters between elite standing Para (two men; one woman) and able- bodied (AB) (three men; four women) XC skiers during a classical skiing race. The data from a global navigation satellite system and inertial measurement unit were integrated to compare time loss and selected sub-techniques as a function of speed. Compared to male/female AB skiers, male/female Para skiers displayed 19/14% slower average speed with the largest time loss (65 ± 36/35 ± 6 s/lap) Citation: Carlsen, C.H.; Kathrin found in uphill terrain. -

Field Indicators of Hydric Soils
United States Department of Field Indicators of Agriculture Natural Resources Hydric Soils in the Conservation Service United States In cooperation with A Guide for Identifying and Delineating the National Technical Committee for Hydric Soils Hydric Soils, Version 8.2, 2018 Field Indicators of Hydric Soils in the United States A Guide for Identifying and Delineating Hydric Soils Version 8.2, 2018 (Including revisions to versions 8.0 and 8.1) United States Department of Agriculture, Natural Resources Conservation Service, in cooperation with the National Technical Committee for Hydric Soils Edited by L.M. Vasilas, Soil Scientist, NRCS, Washington, DC; G.W. Hurt, Soil Scientist, University of Florida, Gainesville, FL; and J.F. Berkowitz, Soil Scientist, USACE, Vicksburg, MS ii In accordance with Federal civil rights law and U.S. Department of Agriculture (USDA) civil rights regulations and policies, the USDA, its Agencies, offices, and employees, and institutions participating in or administering USDA programs are prohibited from discriminating based on race, color, national origin, religion, sex, gender identity (including gender expression), sexual orientation, disability, age, marital status, family/parental status, income derived from a public assistance program, political beliefs, or reprisal or retaliation for prior civil rights activity, in any program or activity conducted or funded by USDA (not all bases apply to all programs). Remedies and complaint filing deadlines vary by program or incident. Persons with disabilities who require alternative means of communication for program information (e.g., Braille, large print, audiotape, American Sign Language, etc.) should contact the responsible Agency or USDA’s TARGET Center at (202) 720-2600 (voice and TTY) or contact USDA through the Federal Relay Service at (800) 877-8339. -

Instrument Rating ‒ Airplane Airman Certification Standards
FAA-S-ACS-8B (with Change 1) U.S. Department of Transportation Federal Aviation Administration Instrument Rating ‒ Airplane Airman Certification Standards June 2018 Flight Standards Service Washington, DC 20591 Acknowledgments The U.S. Department of Transportation, Federal Aviation Administration (FAA), Office of Safety Standards, Regulatory Support Division, Airman Testing Branch, P.O. Box 25082, Oklahoma City, OK 73125 developed this Airman Certification Standards (ACS) document with the assistance of the aviation community. The FAA gratefully acknowledges the valuable support from the many individuals and organizations who contributed their time and expertise to assist in this endeavor. Availability This ACS is available for download from www.faa.gov. Please send comments regarding this document using the following link to the Airman Testing Branch Mailbox. Material in FAA-S-ACS-8B will be effective June 11, 2018. All previous editions of the Instrument Rating – Airplane Airman Certification Standards will be obsolete as of this date for airplane applicants. i Foreword The Federal Aviation Administration (FAA) has published the Instrument Rating – Airplane Airman Certification Standards (ACS) document to communicate the aeronautical knowledge, risk management, and flight proficiency standards for the instrument rating in the airplane category, single-engine land and sea; and multiengine land and sea classes. This ACS incorporates and supersedes FAA-S-ACS-8A Instrument Rating – Airplane Airman Certification Standards. The FAA views the ACS as the foundation of its transition to a more integrated and systematic approach to airman certification. The ACS is part of the Safety Management System (SMS) framework that the FAA uses to mitigate risks associated with airman certification training and testing. -

English Para Swimming Male Short Course Records
ENGLISH PARA SWIMMING MALE SHORT COURSE RECORDS Male 50m Freestyle Name Class Time Meet Venue Date Standard Time S1 02.51.32 01/10/2016 Standard Time S2 01.08.76 01/10/2016 Standard Time S3 00.55.54 01/10/2016 Standard Time S4 00.47.76 01/10/2016 Standard Time S5 00.35.40 01/10/2016 Standard Time S6 00.32.04 01/10/2016 Standard Time S7 00.27.27 01/10/2016 Standard Time S8 00.27.68 01/10/2016 Lewis White S9 00.26.19 National Para-Swimming Championships Manchester 09/12/2017 Standard Time S10 00.24.30 01/10/2016 Standard Time S11 00.29.10 01/10/2016 Standard Time S12 00.25.66 01/10/2016 Standard Time S13 00.25.47 01/10/2016 Thomas Hamer S14 00.24.26 National Para-Swimming Championships Manchester 09/12/2017 Male 100m Freestyle Name Class Time Meet Venue Date Standard Time S1 05.12.29 01/10/2016 Standard Time S2 02.24.62 01/10/2016 Standard Time S3 01.58.04 01/10/2016 Standard Time S4 01.46.85 01/10/2016 Standard Time S5 01.16.23 01/10/2016 Standard Time S6 01.09.21 01/10/2016 Standard Time S7 01.01.03 01/10/2016 Standard Time S8 01.00.22 01/10/2016 Lewis White S9 00.55.71 National Para-Swimming Championships Manchester 09/12/2017 Standard Time S10 00.53.29 01/10/2016 Standard Time S11 01.03.32 01/10/2016 Stephen Clegg S12 00:53.54 National Para-Swimming Championships Manchester 09/12/2017 Standard Time S13 00.55.81 01/10/2016 Reece Dunn S14 00.51.01 SE South West Region Winter Championships 2018 Millfield School 04/11/2018 Male 200m Freestyle Name Class Time Meet Venue Date Standard Time S1 Vacant 01/10/2016 Standard Time S2 05.09.99 01/10/2016 -

Examining Swimming Coaches' Understandings of Inclusion And
Sociology of Sport Journal, 2019, 36, 311-321 https://doi.org/10.1123/ssj.2018-0164 © 2019 Human Kinetics, Inc. ARTICLE “I Feel We are Inclusive Enough”: Examining Swimming Coaches’ Understandings of Inclusion and Disability Andrew Hammond Ruth Jeanes University of British Columbia Monash University, Melbourne Dawn Penney Deana Leahy Edith Cowan University and Monash University, Melbourne Monash University, Melbourne In this study, semi-structured interviews were conducted with eight Victorian swimming coaches to examine the discourses of disability1 and inclusion that they expressed in relation to their current coaching practices. Analysis specifically pursued links between neoliberalism, ableism, elitism, classification and inclusion in coaching, with the intention of exploring what discourse relations are possible, imaginable and practical within what have been referred to as neoliberal-ableist times. Findings reveal that coaches replicate and reproduce elitist, ableist assumptions about the body and sport. The discussion prompts a consideration of how rationalities and techniques of inclusion are limited under the prevailing political context. This paper explores the effects of “neoliberal-able rationality” are central to this research and then expand upon our theorization sport policy and swimming coaches’ understandings of inclusion and application of the concept of neoliberal-ableism to critically and disability. Recent research has highlighted how economic examine sport and inclusion. policies underpinned by neoliberal rationalities of government often see sport as a tool that can be used to alleviate social problems Neoliberalism that are conceived to be a social and economic burden on the state (e.g., juvenile delinquency, obesity, homelessness, unemployment We define neoliberal rationality as a governing rationality through and so on) (Gard, Dionigi, & Dionigi, 2018; Hall, 2006; Parnell, which everything becomes “economized.” As Wendy Brown has May, Widdop, Cope, & Bailey, 2018). -

Rev Bras Cineantropomhum
Rev Bras Cineantropom Hum original article DOI: http://dx.doi.org/10.5007/1980-0037.2017v19n2p196 Sport classification for athletes with visual impairment and its relation with swimming performance Classificação esportiva para atletas com deficiência visual e sua relação com o desempenho na natação Elaine Cappellazzo Souto1,2 Leonardo dos Santos Oliveira1 Claudemir da Silva Santos2 Márcia Greguol1 Abstract – The medical classification (MC) adopted for swimmers with vision visual impairment (VI) does not clearly elucidate the influence of vision loss on performance. In a documentary research, the final time in the 50-, 100- and 400-m freestyle events and MC (S11, S12 and S13) of national (n = 40) and international (n = 72) elite swimmers was analyzed. The analysis was performed using the Kruskal-Wallis test and Spearman’s correlation with 95% confidence (P < 0.05) and Cohen’s d was calculated. There was a large effect of MC on the final time in the 50-m (P = 0.034, d = 1.55) for national ath- letes and in the 50-m (P = 0.001, d = 2.64), 100-m (P = 0.001, d = 3.01) and 400-m (P = 0.001, d = 2.88) for international athletes. S12 and S13 classes were faster compared to S11 class for all international events, but only in the 50-m for the national event (P < 0.05). It was found a strong negative relationship between the final time and MC for international athletes (Spearman’s Rho ≥ 0.78). There was a significant influence of MC on the performance of swimmers in freestyle races, especially in international swimmers. -

Indianapolis 2017 World Para Swimming World S - 6/9/2017 to 6/11/2017 Results - Saturday, June 10 - Finals
USOC - Texas Regionals - Organization License HY-TEK's MEET MANAGER 5.0 - 6:21 PM 6/10/2017 Page 1 Indianapolis 2017 World Para Swimming World S - 6/9/2017 to 6/11/2017 Results - Saturday, June 10 - Finals Event 13 Women 50 LC Meter Backstroke Multi-Class S1 Name Age Team Prelim Time Finals Time Finals 1 Sprenger, Breanna M S1 15 Ridgewood YMCA Dolphins-OH 2:47.79 2:39.04 Event 13 Women 50 LC Meter Backstroke Multi-Class S4 Name Age Team Prelim Time Finals Time Finals 1 Smith, Leanne E S4 29 Usar-CO 1:32.26 1:11.26 2 Cunnington, Tammy S4 41 Canadian Paralympi Team 1:14.02 1:14.27 3 Urhan Giraldo, Angela Victoria S4 45 Colombian Paralympic Team 2:17.23 2:20.42 Event 13 Women 50 LC Meter Backstroke Multi-Class S5 Name Age Team Prelim Time Finals Time Finals 1 Beelner, Vica S5 16 Dynamo Swim Club (Dyna)-GA 1:17.49 1:13.87 2 Serna Cano, Manuela S5 21 Colombian Paralympic Team 1:57.90 1:58.12 Event 14 Men 50 LC Meter Backstroke Multi-Class S2 Name Age Team Prelim Time Finals Time Finals 1 Lovejoy, Curtis S2 60 City of Atlanta Swim Dolphins-GA 1:18.29 1:18.45 2 Lopez Fernandez, Patricio Tse Anibal 36 S2 Dominican Republic Paralympic 1:42.96 1:37.08 Event 14 Men 50 LC Meter Backstroke Multi-Class S3 Name Age Team Prelim Time Finals Time Finals 1 Lopez Diaz, Diego S3 22 Mexico Paralymic Team 1:03.16 1:08.95 Event 14 Men 50 LC Meter Backstroke Multi-Class S4 Name Age Team Prelim Time Finals Time Finals 1 McLean, Riley S4 17 Canadian Paralympi Team 57.98 56.84 2 Zarate Rodriguez, Marcos Rafael S4 17 Mexico Paralymic Team 1:06.10 1:03.46 3 Marsh, -
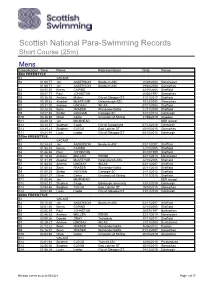
Para Swimming Records Short Course As At
Scottish National Para-Swimming Records Short Course (25m) Mens Classification Time Name Representation Date Venue 50m FREESTYLE S1 VACANT S2 01:08.77 Jim ANDERSON Broxburn ASC 26/08/2004 Manchester 01:08.77 Jim ANDERSON Broxburn ASC 19/04/2008 Glenrothes S3 00:55.55 Kenny CAIRNS 23/10/2005 Sheffield S4 00:47.77 Paul JOHNSTON 26/04/1997 Glenrothes S5 00:36.06 Andrew Mullen City of Glasgow ST 11/11/2017 Sheffield S6 00:39.82 Alasdair McARTHUR Helensburgh ASC 10/12/2005 Glenrothes S7 00:31.20 Andrew LINDSAY INCAS 07/11/2004 Sheffield S8 00:28.08 Sean FRASER Warrender Baths 22/11/2009 Sheffield S9 00:27.58 Stefan HOGGAN Carnegie SC 13/12/2013 Edinburgh S10 00:26.59 Oliver Carter University of Stirling 21/09/2019 Glasgow S11 00:29.74 Jim MUIRHEAD BBS record S12 00:24.37 Stephen Clegg City of Sunderland 07/12/2018 Edinburgh S13 00:25.23 Stephen CLEGG East Lothian ST 25/10/2015 Glenrothes S14 00:24.94 Louis Lawlor City of Glasgow ST 08/12/2018 Edinburgh 100m FREESTYLE S1 VACANT S2 02:24.63 Jim ANDERSON Broxburn ASC 03/11/2007 Sheffield S3 01:58.05 Kenny CAIRNS 22/10/2005 Sheffield S4 01:46.86 Paul JOHNSTON 30/10/1999 Sheffield S5 01:18.26 Andrew MULLEN REN96 22/11/2014 Manchester S6 01:31.89 Alasdair McARTHUR Helensburgh ASC 22/10/2005 Sheffield S7 01:08.00 Andrew LINDSAY INCAS 03/11/2007 Sheffield S8 01:00.64 Sean FRASER Warrender Baths 20/11/2010 Sheffield S9 01:00.35 Stefan HOGGAN Carnegie SC 24/11/2012 Sheffield S10 00:57.27 Oliver Carter University of Stirling 11/11/2018 Sheffield S11 01:05.46 Jim MUIRHEAD BBS record S12 00:52.31 Stephen