Genetic Risk of Autism Spectrum Disorder in a Pakistani Population
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Learning from Cadherin Structures and Sequences: Affinity Determinants and Protein Architecture
Learning from cadherin structures and sequences: affinity determinants and protein architecture Klára Fels ıvályi Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Klara Felsovalyi All rights reserved ABSTRACT Learning from cadherin structures and sequences: affinity determinants and protein architecture Klara Felsovalyi Cadherins are a family of cell-surface proteins mediating adhesion that are important in development and maintenance of tissues. The family is defined by the repeating cadherin domain (EC) in their extracellular region, but they are diverse in terms of protein size, architecture and cellular function. The best-understood subfamily is the type I classical cadherins, which are found in vertebrates and have five EC domains. Among the five different type I classical cadherins, the binding interactions are highly specific in their homo- and heterophilic binding affinities, though their sequences are very similar. As previously shown, E- and N-cadherins, two prototypic members of the subfamily, differ in their homophilic K D by about an order of magnitude, while their heterophilic affinity is intermediate. To examine the source of the binding affinity differences among type I cadherins, we used crystal structures, analytical ultracentrifugation (AUC), surface plasmon resonance (SPR), and electron paramagnetic resonance (EPR) studies. Phylogenetic analysis and binding affinity behavior show that the type I cadherins can be further divided into two subgroups, with E- and N-cadherin representing each. In addition to the affinity differences in their wild-type binding through the strand-swapped interface, a second interface also shows an affinity difference between E- and N-cadherin. -

Somatic Mutational Landscapes of Adherens Junctions and Their
1 Somatic mutational landscapes of adherens junctions and their 2 functional consequences in cutaneous melanoma development 3 4 Praveen Kumar Korla,1 Chih-Chieh Chen,2 Daniel Esguerra Gracilla,1 Ming-Tsung Lai,3 Chih- 5 Mei Chen,4 Huan Yuan Chen,5 Tritium Hwang,1 Shih-Yin Chen,4,6,* Jim Jinn-Chyuan Sheu1,4, 6-9,* 6 1Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan; 7 2Institute of Medical Science and Technology, National Sun Yat-sen University, Kaohsiung 80424, 8 Taiwan; 3Department of Pathology, Taichung Hospital, Ministry of Health and Welfare, Taichung 9 40343, Taiwan; 4Genetics Center, China Medical University Hospital, Taichung 40447, Taiwan; 10 5Institute of Biomedical Sciences, Academia Sinica, Taipei 11574, Taiwan; 6School of Chinese 11 Medicine, China Medical University, Taichung 40402, Taiwan; 7Department of Health and 12 Nutrition Biotechnology, Asia University, Taichung 41354, Taiwan; 8Department of 13 Biotechnology, Kaohsiung Medical University, Kaohsiung 80708, Taiwan; 9Institute of 14 Biopharmaceutical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan 15 16 PKK, CCC and DEG contributed equally to this study. 17 *Correspondence to: Dr. Shih-Yin Chen ([email protected]) at Genetics Center, China 18 Medical University Hospital, Taichung, 40447, TAIWAN; or Dr. Jim Jinn-Chyuan Sheu 19 ([email protected]) at Institute of Biomedical Sciences, National Sun Yat-sen 20 University, Kaohsiung City 80424, TAIWAN. 21 22 Running title: mutational landscape of cadherins in melanoma 1 23 Abstract 24 Cell-cell interaction in skin homeostasis is tightly controlled by adherens junctions (AJs). 25 Alterations in such regulation lead to melanoma development. -

Pathway and Network Analysis of Somatic Mutations Across Cancer
Network Analysis of Mutaons Across Cancer Types Ben Raphael Fabio Vandin, Max Leiserson, Hsin-Ta Wu Department of Computer Science Center for Computaonal Molecular Biology Significantly Mutated Genes Muta#on Matrix Stascal test Genes Paents Frequency Number Paents Study Num. Samples Num. SMG TCGA Ovarian (2011) 316 10 TCGA Breast (2012) 510 35 TCGA Colorectal (2012) 276 32 background mutaon rate (BMR), gene specific effects, etc. Significantly Mutated Genes à Pathways Stascal test Frequency Number Paents TCGA Colorectal (Nature 2012) TCGA Ovarian (Nature 2011) background mutaon rate (BMR), gene specific effects, etc. Advantages of Large Datasets Prior knowledge of groups of genes Genes Paents Known pathways Interac3on Network None Prior knowledge • Novel pathways or interac3ons between pathways (crosstalk) • Topology of interac3ons Two Algorithms Prior knowledge of groups of genes Genes Paents Known pathways Interac3on Network None Prior knowledge Number of Hypotheses HotNet subnetworks of Dendrix interac3on network Exclusive gene sets HotNet: Problem Defini3on Given: 1. Network G = (V, E) V = genes. E = interac3ons b/w genes 2. Binary mutaon matrix Genes = mutated = not mutated Paents Find: Connected subnetworks mutated in a significant number of paents. Subnetwork Properes Mutaon frequency/score AND network topology Frequency Number Paents • Moderate frequency/score • High frequency/score • Highly connected • Connected through high-degree node. Example: TP53 has 238 neighbors in HPRD network Mutated subnetworks: HotNet* Muta#on Matrix Human Interac#on Network Genes = mutated genes Paents (1) Muta#on à heat diffusion Extract “significantly hot” subnetworks Hot (2) Cold *F. Vandin, E. Upfal, and B. J. Raphael. J. Comp.Biol. (2011). Also RECOMB (2010). Stas3cal Test Muta#on Matrix Random Binary Matrix Genes Genes Paents Paents Xs = number of subnetworks ≥ s genes Two-stage mul-hypothesis test: Rigorously bound FDR. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

The Untold Stories of the Speech Gene, the FOXP2 Cancer Gene
www.Genes&Cancer.com Genes & Cancer, Vol. 9 (1-2), January 2018 The untold stories of the speech gene, the FOXP2 cancer gene Maria Jesus Herrero1,* and Yorick Gitton2,* 1 Center for Neuroscience Research, Children’s National Medical Center, NW, Washington, DC, USA 2 Sorbonne University, INSERM, CNRS, Vision Institute Research Center, Paris, France * Both authors contributed equally to this work Correspondence to: Yorick Gitton, email: [email protected] Keywords: FOXP2 factor, oncogene, cancer, prognosis, language Received: March 01, 2018 Accepted: April 02, 2018 Published: April 18, 2018 Copyright: Herrero and Gitton et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT FOXP2 encodes a transcription factor involved in speech and language acquisition. Growing evidence now suggests that dysregulated FOXP2 activity may also be instrumental in human oncogenesis, along the lines of other cardinal developmental transcription factors such as DLX5 and DLX6 [1–4]. Several FOXP family members are directly involved during cancer initiation, maintenance and progression in the adult [5–8]. This may comprise either a pro- oncogenic activity or a deficient tumor-suppressor role, depending upon cell types and associated signaling pathways. While FOXP2 is expressed in numerous cell types, its expression has been found to be down-regulated in breast cancer [9], hepatocellular carcinoma [8] and gastric cancer biopsies [10]. Conversely, overexpressed FOXP2 has been reported in multiple myelomas, MGUS (Monoclonal Gammopathy of Undetermined Significance), several subtypes of lymphomas [5,11], as well as in neuroblastomas [12] and ERG fusion-negative prostate cancers [13]. -
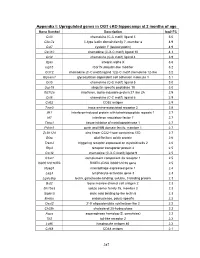
PDF (Appendices)
Appendix I: Upregulated genes in OGT cKO hippocampi at 2 months of age Gene Symbol Description log2 FC Ccl3 chemokine (C-C motif) ligand 3 5.0 Clec7a C-type lectin domain family 7, member a 4.9 Cst7 cystatin F (leukocystatin) 4.9 Cxcl10 chemokine (C-X-C motif) ligand 10 4.3 Ccl4 chemokine (C-C motif) ligand 4 3.9 Itgax integrin alpha X 3.6 Isg15 ISG15 ubiquitin-like modifier 3.2 Ccl12 chemokine (C-C motif) ligand 12|c-C motif chemokine 12-like 3.2 Glycam1 glycosylation dependent cell adhesion molecule 1 3.1 Ccl5 chemokine (C-C motif) ligand 5 3.0 Usp18 ubiquitin specific peptidase 18 3.0 Ifi27l2a interferon, alpha-inducible protein 27 like 2A 2.9 Ccl6 chemokine (C-C motif) ligand 6 2.9 Cd52 CD52 antigen 2.9 Taar3 trace amine-associated receptor 3 2.8 Ifit1 interferon-induced protein with tetratricopeptide repeats 1 2.7 Irf7 interferon regulatory factor 7 2.7 Timp1 tissue inhibitor of metalloproteinase 1 2.7 Pyhin1 pyrin and HIN domain family, member 1 2.7 Zc3h12d zinc finger CCCH type containing 12D 2.7 Gfap glial fibrillary acidic protein 2.6 Trem2 triggering receptor expressed on myeloid cells 2 2.6 Rtp4 receptor transporter protein 4 2.5 Cxcl9 chemokine (C-X-C motif) ligand 9 2.5 C3ar1 complement component 3a receptor 1 2.5 I830012O16Rik RIKEN cDNA I830012O16 gene 2.5 Mpeg1 macrophage expressed gene 1 2.4 Lag3 lymphocyte-activation gene 3 2.3 Lgals3bp lectin, galactoside-binding, soluble, 3 binding protein 2.3 Bst2 bone marrow stromal cell antigen 2 2.3 Slc15a3 solute carrier family 15, member 3 2.3 Siglec5 sialic acid binding Ig-like lectin 5 2.3 Endou endonuclease, polyU-specific 2.2 Oasl2 2'-5' oligoadenylate synthetase-like 2 2.2 Ch25h cholesterol 25-hydroxylase 2.2 Aspg asparaginase homolog (S. -

Chr CNV Start CNV Stop Gene Gene Feature 1 37261312 37269719
chr CNV start CNV stop Gene Gene feature 1 37261312 37269719 Tmem131 closest upstream gene 1 37261312 37269719 Cnga3 closest downstream gene 1 41160869 41180390 Tmem182 closest upstream gene 1 41160869 41180390 2610017I09Rik closest downstream gene 1 66835123 66839616 1110028C15Rik in region 2 88714200 88719211 Olfr1206 closest upstream gene 2 88714200 88719211 Olfr1208 closest downstream gene 2 154840037 154846228 a in region 3 30065831 30417157 Mecom closest upstream gene 3 30065831 30417157 Arpm1 closest downstream gene 3 35476875 35495913 Sox2ot closest upstream gene 3 35476875 35495913 Atp11b closest downstream gene 3 39563408 39598697 Fat4 closest upstream gene 3 39563408 39598697 Intu closest downstream gene 3 94246481 94410611 Celf3 in region 3 94246481 94410611 Mrpl9 in region 3 94246481 94410611 Riiad1 in region 3 94246481 94410611 Snx27 in region 3 104311901 104319916 Lrig2 in region 3 144613709 144619149 Clca6 in region 3 144613709 144619149 Clca6 in region 4 108673 137301 Vmn1r2 closest downstream gene 4 3353037 5882883 6330407A03Rik in region 4 3353037 5882883 Chchd7 in region 4 3353037 5882883 Fam110b in region 4 3353037 5882883 Impad1 in region 4 3353037 5882883 Lyn in region 4 3353037 5882883 Mos in region 4 3353037 5882883 Penk in region 4 3353037 5882883 Plag1 in region 4 3353037 5882883 Rps20 in region 4 3353037 5882883 Sdr16c5 in region 4 3353037 5882883 Sdr16c6 in region 4 3353037 5882883 Tgs1 in region 4 3353037 5882883 Tmem68 in region 4 5919294 6304249 Cyp7a1 in region 4 5919294 6304249 Sdcbp in region 4 5919294 -

The Central Role of Cadherins in Gonad Development, Reproduction and Fertility
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 2 October 2020 doi:10.20944/preprints202010.0037.v1 Review The central role of cadherins in gonad development, reproduction and fertility Rafal P. Piprek 1,*, Malgorzata Kloc 2,3,4, Paulina Mizia 1 and Jacek Z. Kubiak 5,6,* 1 Department of Comparative Anatomy, Institute of Zoology and Biomedical Research, Jagiellonian University, Gronostajowa 9, 30-387 Krakow, Poland; e-mail [email protected] (R.P.P.), [email protected] (P.M.) 2 The Houston Methodist Research Institute, Houston, TX 77030, USA; e-mail [email protected] 3 Department of Surgery, The Houston Methodist Hospital, Houston, TX 77030, USA 4 MD Anderson Cancer Center, University of Texas, Houston, TX 77030, USA 5 UnivRennes, UMR 6290 CNRS/UR1, Institute of Genetics and Development of Rennes, Cycle Group, Faculty of Medicine, F-35000 Rennes, France; e-mail [email protected] 6 Department of Regenerative Medicine and Cell Biology, Military Institute of Hygiene and Epidemiology (WIHE), 01-163 Warsaw, Poland * Correspondence: [email protected]; +330612253086 (J.Z.K) [email protected]; Tel.: +48126645059 (R.P.P.) Abstract: Cadherins are a group of membrane proteins responsible for cell adhesion. They are crucial for cell sorting and recognition during the morphogenesis, but also play many other roles such as assuring tissue integrity and resistance to stretching, mechanotransduction, cell signaling, regulation of cell proliferation, apoptosis, survival, carcinogenesis, etc. Within the cadherin superfamily, the E- and N-cadherin have been especially well studied. They are involved in many aspects of sexual development and reproduction, such as germline development and gametogenesis, gonad development and functioning, and fertilization. -

Sequential Changes in Gene Expression Profiles in Breast Cancers During Treatment with the Aromatase Inhibitor, Letrozole
The Pharmacogenomics Journal (2012) 12, 10–21 & 2012 Macmillan Publishers Limited. All rights reserved 1470-269X/12 www.nature.com/tpj ORIGINAL ARTICLE Sequential changes in gene expression profiles in breast cancers during treatment with the aromatase inhibitor, letrozole WR Miller1, A Larionov1, The study aim was to identify early (within 14 days) and late changes (by 3 1 2 months) in breast cancer gene expression profiles associated with neoadju- TJ Anderson , DB Evans and vant therapy with letrozole. RNA from sequential tumour biopsies in 54 1 JM Dixon patients was analyzed on microarrays; changes were determined by frequency, magnitude and significance analyses. Substantially more genes 1Edinburgh Breast Unit Research Group, Western General Hospital, Edinburgh, UK and 2Novartis were changed at 3 months (1503) than at 14 days (237). Early changed Institutes for Biomedical Research Basel, Oncology genes were associated with cell cycle (downregulation), blood vessel Research, Basel, Switzerland development and extracellular matrix (upregulation); late changes included ‘cellular metabolic process’, ‘generation of precursor metabolites and energy’ Correspondence: (decreased) and ‘cell adhesion’ ‘biological adhesion‘ (increased). A striking Professor WR Miller, 2 Stoneycroft Road, South Queensferry EH30 9HX, UK. difference between the early and late changes was the general location of E-mail: [email protected] downregulated genes—nuclear structures at 14 days and mitochondria after 3 months. These changes in gene expression profiles -

RNA-Seq Data Analysis of a Rodent Model of Adolescent Binge Drinking Reveals Pathways
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.02.365841; this version posted November 3, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. RNA-Seq data analysis of a rodent model of adolescent binge drinking reveals pathways and candidate genes involved in neuronal remodeling and neuroimmune activation Alejandro Q. Nato, Jr.1,†, Hafiz Ata Ul Mustafa1*, Riya K. Patel1*, Hannah G. Sexton1,2, Scott D. Moore3,4, James Denvir1, Donald A. Primerano1, and Mary-Louise Risher1,2,3,† 1Department of Biomedical Sciences, Joan C. Edwards School of Medicine, Marshall University, Huntington, WV, USA 2Hershel ‘Woody’ Williams Veteran Affairs Medical Center, Huntington WV, USA 3Department of Psychiatry and Behavioral Sciences, Duke University Medical Center, Durham, NC, USA 4Durham Veteran Affairs Medical Center, Durham, NC, USA *equal contribution †Addresses for correspondence: Alejandro Q. Nato, Jr., 1700 Third Ave (BBSC 336M), Huntington, WV 25755 Tel: 304 6963562 Fax: 304 6967207 Email: [email protected] Mary-Louise Risher, 1700 Third Ave (BBSC 336L), Huntington, WV 25755 Tel: 304 6963894 Fax: 304 6967207 Email: [email protected] Running titles: Identification of candidate genes for alcohol use disorder using RNA-seq data; Pathway functional and network enrichment analyses of RNA-Seq data in alcohol use disorder bioRxiv preprint doi: https://doi.org/10.1101/2020.11.02.365841; this version posted November 3, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. -

Blood Biomarkers for Memory: Toward Early Detection of Risk for Alzheimer Disease, Pharmacogenomics, and Repurposed Drugs
Molecular Psychiatry (2020) 25:1651–1672 https://doi.org/10.1038/s41380-019-0602-2 IMMEDIATE COMMUNICATION Blood biomarkers for memory: toward early detection of risk for Alzheimer disease, pharmacogenomics, and repurposed drugs 1,2,3 1 1 1,3,4 1 1 1 3 A. B. Niculescu ● H. Le-Niculescu ● K. Roseberry ● S. Wang ● J. Hart ● A. Kaur ● H. Robertson ● T. Jones ● 3 3,5 5 2 1 1,4 4 A. Strasburger ● A. Williams ● S. M. Kurian ● B. Lamb ● A. Shekhar ● D. K. Lahiri ● A. J. Saykin Received: 25 March 2019 / Revised: 25 September 2019 / Accepted: 11 November 2019 / Published online: 2 December 2019 © The Author(s) 2019. This article is published with open access Abstract Short-term memory dysfunction is a key early feature of Alzheimer’s disease (AD). Psychiatric patients may be at higher risk for memory dysfunction and subsequent AD due to the negative effects of stress and depression on the brain. We carried out longitudinal within-subject studies in male and female psychiatric patients to discover blood gene expression biomarkers that track short term memory as measured by the retention measure in the Hopkins Verbal Learning Test. These biomarkers were subsequently prioritized with a convergent functional genomics approach using previous evidence in the field implicating them in AD. The top candidate biomarkers were then tested in an independent cohort for ability to predict state short-term memory, 1234567890();,: 1234567890();,: and trait future positive neuropsychological testing for cognitive impairment. The best overall evidence was for a series of new, as well as some previously known genes, which are now newly shown to have functional evidence in humans as blood biomarkers: RAB7A, NPC2, TGFB1, GAP43, ARSB, PER1, GUSB, and MAPT. -

CNTNAP2 Gene in High Functioning Autism
CNTNAP2 gene in high functioning autism UCCAP Anna Werling 1, Regina Taurines 4, Elise Bobrowski 1,3 , Ronnie Gundelfinger 1, Marcel Romanos 4, Edna Grünblatt 1,2 , Susanne Walitza 1,2 1 University Clinics of Child and Adolescent Psychiatry, University of Zurich, Switzerland; 2 Neuroscience Center Zurich, University of Zurich and ETH Zurich, Switzerland Zurich 3 Department of Experimental Psychology, University of Regensburg, Regensburg, Germany; 4 Department of Child and Adolescent Psychiatry, Psychosomatics and Psychotherapy, University of Würzburg, Germany Introduction: Autism spectrum disorder (ASD) is a neurodevelopmental disorder collectively characterized by the trias of impairments in social interaction and communication and by repetitive or unusual behaviors. ASDs include early infantile Autism, Asperger’s disorder and atypical Autism. ASD is often associated with intellectual disability and speech/language impairments. According to multiple family and twin studies with a substantial heritable component with 70-90% of autism etiology, ASD is considered to be one of he most strongly genetically influenced multifactorial childhood psychiatric disorder. Despite the high heritability, it is interfered to be complex and no major gene has been observed to be relevant for the majority of ASD diagnoses. In the past decades various studies have implicated a small amount of genes with either rare highly-penetrant mutations, low-penetrant common variants or copy-number variants (CNV) that together explain only about 15-30% of the population prevalence. The Contactin Associated Protein-like 2 (CNTNAP2) gene has been discussed to be associated in ASD and other neurodevelopmental disorders. In this study, we aimed to elucidate the genetic association of CNTNAP2 gene within high functioning ASD (HFA) to eliminate intelligence related factors and combine all published results to-date in a meta-analysis for ASD and HFA .