Accelerated Regulatory Gene Evolution in an Adaptive Radiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PRIORITY GRASSLANDS INITIATIVE Methodology for Identifying Priority Grasslands
PRIORITY GRASSLANDS INITIATIVE Methodology for Identifying Priority Grasslands September 2007 Building a Scientific Framework and Rationale for Sustainable Conservation and Stewardship Grasslands Conservation Council of British Columbia 954A Laval Crescent Kamloops, BC V2C 5P5 Phone: (250) 374-5787 Email: [email protected] Website: www.bcgrasslands.org Cover photo: Prickly-pear cactus by Richard Doucette Grasslands Conservation Council of British Columbia i Table of Contents Executive Summary .....................................................................................................................................i Acknowledgements .....................................................................................................................................ii Introduction.................................................................................................................................................1 Methodology Overview...............................................................................................................................2 Methodology Stages ....................................................................................................................................5 Stage 1: Initial GIS Data Gathering, Preparation and Analysis................................................................ 5 Stage 2: Expert Input .............................................................................................................................. 10 Stage 3: Assessment of Recreational -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Chapter 1 the California Flora
CHAPTER 1 THE CALIFORNIA FLORA The Californian Floristic Province California is a large state with a complex topography and a great diversity of climates and habitats,resulting in a very large assemblage of plant species that vary in size and include both the world’s largest trees and some of the smallest and most unique plant species. In order to create manageable units for plant investigations, botanists have divided the continental landform into geographic units called floristic provinces. These units reflect the wide variations in natural landscapes and assist botanists in predicting where a given plant might be found. Within the borders of California, there are three floristic provinces, each extending beyond the state’s political boundaries. The California Floristic Province includes the geographi- cal area that contains assemblages of plant species that are more or less characteristic of California and that are best de- veloped in the state.This province includes southwestern Ore- gon and northern Baja California but excludes certain areas of the southeastern California desert regions, as well as the area of the state that is east of the Sierra Nevada–Cascade Range axis (map 1).The flora of the desert areas and those east of the Sierra Nevada crest are best developed outside the state, and therefore, parts of the state of California are not in the Cali- fornia Floristic Province. The Great Basin Floristic Province includes some of the area east of the Sierra Nevada and some regions in the northeastern part of the state, although some botanists consider the latter area to belong to another distinct floristic province, the Columbia Plateau Floristic Province. -

Breeding System Diversification and Evolution in American Poa Supersect. Homalopoa (Poaceae: Poeae: Poinae)
Annals of Botany Page 1 of 23 doi:10.1093/aob/mcw108, available online at www.aob.oxfordjournals.org Breeding system diversification and evolution in American Poa supersect. Homalopoa (Poaceae: Poeae: Poinae) Liliana M. Giussani1,*, Lynn J. Gillespie2, M. Amalia Scataglini1,Marıa A. Negritto3, Ana M. Anton4 and Robert J. Soreng5 1Instituto de Botanica Darwinion, San Isidro, Buenos Aires, Argentina, 2Research and Collections Division, Canadian Museum of Nature, Ottawa, Ontario, Canada, 3Universidad de Magdalena, Santa Marta, Colombia, 4Instituto Multidisciplinario de Biologıa Vegetal (IMBIV), CONICET-UNC, Cordoba, Argentina and 5Department of Botany, Smithsonian Institution, Washington, DC, USA *For correspondence. E-mail [email protected] Received: 11 December 2015 Returned for revision: 18 February 2016 Accepted: 18 March 2016 Downloaded from Background and Aims Poa subgenus Poa supersect. Homalopoa has diversified extensively in the Americas. Over half of the species in the supersection are diclinous; most of these are from the New World, while a few are from South-East Asia. Diclinism in Homalopoa can be divided into three main types: gynomonoecism, gynodioe- cism and dioecism. Here the sampling of species of New World Homalopoa is expanded to date its origin and diver- sification in North and South America and examine the evolution and origin of the breeding system diversity. Methods A total of 124 specimens were included in the matrix, of which 89 are species of Poa supersect. http://aob.oxfordjournals.org/ Homalopoa sections Acutifoliae, Anthochloa, Brizoides, Dasypoa, Dioicopoa, Dissanthelium, Homalopoa sensu lato (s.l.), Madropoa and Tovarochloa, and the informal Punapoa group. Bayesian and parsimony analyses were conducted on the data sets based on four markers: the nuclear ribosomal internal tanscribed spacer (ITS) and exter- nal transcribed spacer (ETS), and plastid trnT-L and trnL-F. -

Media Emerging
The Bureau of International Information Programs of GLOBAL ISSUES the U.S. Department of State publishes five electronic journals under the eJournal USA logo—Economic Perspectives, Global Issues, Issues of Democracy, Foreign Policy Agenda, and Society Editor Virginia L. Farris & Values—that examine major issues facing the United Managing Editor Charlene Porter States and the international community, as well as U.S. Contributing Editors Jenifer Bochner society, values, thought, and institutions. Kara Breissinger Rhobyn Costen-Sykes One new journal is published monthly in English and Eileen Kane followed by versions in French, Portuguese, Russian, Cynthia LaCovey and Spanish. Selected editions also appear in Arabic Cheryl Pellerin and Chinese. Each journal is catalogued by volume (the Gerri Williams number of years in publication) and number (the number Associate Editor Rosalie Targonski of issues that appear during the year). Reference Specialists Lynne Scheib Joan R. Taylor Opinions expressed in the journals do not necessarily Art Director Tim Brown reflect the views or policies of the U.S. government. The U.S. Department of State assumes no responsibility for Photo Researcher Maggie Johnson Sliker the content and continued accessibility of Internet sites to which the journals link; such responsibility resides solely with the publishers of those sites. Journal articles, Publisher Judith S. Siegel photographs, and illustrations may be reproduced and Senior Editor George Clack translated outside the United States unless they carry Executive Editor Richard W. Huckaby explicit copyright restrictions, in which case permission Production Manager Christian Larson must be sought from the copyright holders noted in the journal. Assistant Production Manager Chloe D. -

Chromosome Numbers in Compositae, XII: Heliantheae
SMITHSONIAN CONTRIBUTIONS TO BOTANY 0 NCTMBER 52 Chromosome Numbers in Compositae, XII: Heliantheae Harold Robinson, A. Michael Powell, Robert M. King, andJames F. Weedin SMITHSONIAN INSTITUTION PRESS City of Washington 1981 ABSTRACT Robinson, Harold, A. Michael Powell, Robert M. King, and James F. Weedin. Chromosome Numbers in Compositae, XII: Heliantheae. Smithsonian Contri- butions to Botany, number 52, 28 pages, 3 tables, 1981.-Chromosome reports are provided for 145 populations, including first reports for 33 species and three genera, Garcilassa, Riencourtia, and Helianthopsis. Chromosome numbers are arranged according to Robinson’s recently broadened concept of the Heliantheae, with citations for 212 of the ca. 265 genera and 32 of the 35 subtribes. Diverse elements, including the Ambrosieae, typical Heliantheae, most Helenieae, the Tegeteae, and genera such as Arnica from the Senecioneae, are seen to share a specialized cytological history involving polyploid ancestry. The authors disagree with one another regarding the point at which such polyploidy occurred and on whether subtribes lacking higher numbers, such as the Galinsoginae, share the polyploid ancestry. Numerous examples of aneuploid decrease, secondary polyploidy, and some secondary aneuploid decreases are cited. The Marshalliinae are considered remote from other subtribes and close to the Inuleae. Evidence from related tribes favors an ultimate base of X = 10 for the Heliantheae and at least the subfamily As teroideae. OFFICIALPUBLICATION DATE is handstamped in a limited number of initial copies and is recorded in the Institution’s annual report, Smithsonian Year. SERIESCOVER DESIGN: Leaf clearing from the katsura tree Cercidiphyllumjaponicum Siebold and Zuccarini. Library of Congress Cataloging in Publication Data Main entry under title: Chromosome numbers in Compositae, XII. -
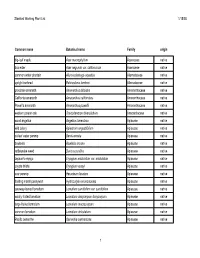
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Download The
SYSTEMATICA OF ARNICA, SUBGENUS AUSTROMONTANA AND A NEW SUBGENUS, CALARNICA (ASTERACEAE:SENECIONEAE) by GERALD BANE STRALEY B.Sc, Virginia Polytechnic Institute, 1968 M.Sc, Ohio University, 1974 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS OF THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES (Department of Botany) We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA March 1980 © Gerald Bane Straley, 1980 In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the Head of my Department or by his representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department nf Botany The University of British Columbia 2075 Wesbrook Place Vancouver, Canada V6T 1W5 26 March 1980 ABSTRACT Seven species are recognized in Arnica subgenus Austromontana and two species in a new subgenus Calarnica based on a critical review and conserva• tive revision of the species. Chromosome numbers are given for 91 populations representing all species, including the first reports for Arnica nevadensis. Results of apomixis, vegetative reproduction, breeding studies, and artifi• cial hybridizations are given. Interrelationships of insect pollinators, leaf miners, achene feeders, and floret feeders are presented. Arnica cordifolia, the ancestral species consists largely of tetraploid populations, which are either autonomous or pseudogamous apomicts, and to a lesser degree diploid, triploid, pentaploid, and hexaploid populations. -

Chloroplast DNA Evidence for a North American Origin of the Hawaiian
Proc, Natl. Acad. Sci. USA Vol. 88, pp. 1840-1843, March 1991 Evolution Chloroplast DNA evidence for a North American origin of the Hawaiian silversword alliance (Asteraceae) (insular evolution/adaptive radiation/biogeography/long-distance dispersal/phylogenetics) BRUCE G. BALDWIN*t, DONALD W. KYHOS*, JAN DVORAKO, AND GERALD D. CARR§ Departments of *Botany and tAgronomy and Range Science, University of California, Davis, CA 95616; and §Department of Botany, University of Hawaii, Honolulu, HI 96822 Communicated by Peter H. Raven, November 30, 1990 (received for review August 1, 1990) ABSTRACT Chloroplast DNA restriction-site compari- sity, structural (4, 5), biosystematic (6), allozymic (7), and sons were made among 24 species of the Hawaiian silversword chloroplast DNA (cpDNA) (8) data indicate the silversword alliance (Argyroxiphium, Dubautia, and Wilkesia) and 7 species alliance originated from a single colonizing species. of North American perennial tarweeds in Adenothamnus, Ma- Carlquist (1) presented convincing anatomical evidence dia, Raillardella, and Railkrdiopsis (Asteraceae-Madiinae). indicating taxonomic alignment of the Hawaiian silversword These data and results from intergeneric hybridization indi- alliance with the almost exclusively herbaceous American cated surprisingly close genetic affinity of the monophyletic Madiinae or tarweeds. Gray (9) earlier suggested such affinity Hawaiian group to two diploid species of montane perennial for Argyroxiphium, which was disputed by Keck (10) based herbs in California, Madia bolanderi and Raillardiopsis muiri. on presumed morphological dissimilarities and the magnitude Of 117 restriction-site mutations shared among a subset of two of the oceanic barrier to migration. Herein, we compare or more accessions, more than one-fifth (25 mutations) sepa- cpDNA restriction sites between the silversword alliance and rated the silversword alliance, M. -

USDA Forest Service, Pacific Southwest Region Sensitive Plant Species by Forest
USDA Forest Service, Pacific Southwest Region 1 Sensitive Plant Species by Forest 2013 FS R5 RF Plant Species List Klamath NF Mendocino NF Shasta-Trinity NF NF Rivers Six Lassen NF Modoc NF Plumas NF EldoradoNF Inyo NF LTBMU Tahoe NF Sequoia NF Sierra NF Stanislaus NF Angeles NF Cleveland NF Los Padres NF San Bernardino NF Scientific Name (Common Name) Abies bracteata (Santa Lucia fir) X Abronia alpina (alpine sand verbena) X Abronia nana ssp. covillei (Coville's dwarf abronia) X X Abronia villosa var. aurita (chaparral sand verbena) X X Acanthoscyphus parishii var. abramsii (Abrams' flowery puncturebract) X X Acanthoscyphus parishii var. cienegensis (Cienega Seca flowery puncturebract) X Agrostis hooveri (Hoover's bentgrass) X Allium hickmanii (Hickman's onion) X Allium howellii var. clokeyi (Mt. Pinos onion) X Allium jepsonii (Jepson's onion) X X Allium marvinii (Yucaipa onion) X Allium tribracteatum (three-bracted onion) X X Allium yosemitense (Yosemite onion) X X Anisocarpus scabridus (scabrid alpine tarplant) X X X Antennaria marginata (white-margined everlasting) X Antirrhinum subcordatum (dimorphic snapdragon) X Arabis rigidissima var. demota (Carson Range rock cress) X X Arctostaphylos cruzensis (Arroyo de la Cruz manzanita) X Arctostaphylos edmundsii (Little Sur manzanita) X Arctostaphylos glandulosa ssp. gabrielensis (San Gabriel manzanita) X X Arctostaphylos hooveri (Hoover's manzanita) X Arctostaphylos luciana (Santa Lucia manzanita) X Arctostaphylos nissenana (Nissenan manzanita) X X Arctostaphylos obispoensis (Bishop manzanita) X Arctostphylos parryana subsp. tumescens (interior manzanita) X X Arctostaphylos pilosula (Santa Margarita manzanita) X Arctostaphylos rainbowensis (rainbow manzanita) X Arctostaphylos refugioensis (Refugio manzanita) X Arenaria lanuginosa ssp. saxosa (rock sandwort) X Astragalus anxius (Ash Valley milk-vetch) X Astragalus bernardinus (San Bernardino milk-vetch) X Astragalus bicristatus (crested milk-vetch) X X Pacific Southwest Region, Regional Forester's Sensitive Species List. -

Edible Seeds and Grains of California Tribes
National Plant Data Team August 2012 Edible Seeds and Grains of California Tribes and the Klamath Tribe of Oregon in the Phoebe Apperson Hearst Museum of Anthropology Collections, University of California, Berkeley August 2012 Cover photos: Left: Maidu woman harvesting tarweed seeds. Courtesy, The Field Museum, CSA1835 Right: Thick patch of elegant madia (Madia elegans) in a blue oak woodland in the Sierra foothills The U.S. Department of Agriculture (USDA) prohibits discrimination in all its pro- grams and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sex- ual orientation, genetic information, political beliefs, reprisal, or because all or a part of an individual’s income is derived from any public assistance program. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA’s TARGET Center at (202) 720-2600 (voice and TDD). To file a complaint of discrimination, write to USDA, Director, Office of Civil Rights, 1400 Independence Avenue, SW., Washington, DC 20250–9410, or call (800) 795-3272 (voice) or (202) 720-6382 (TDD). USDA is an equal opportunity provider and employer. Acknowledgments This report was authored by M. Kat Anderson, ethnoecologist, U.S. Department of Agriculture, Natural Resources Conservation Service (NRCS) and Jim Effenberger, Don Joley, and Deborah J. Lionakis Meyer, senior seed bota- nists, California Department of Food and Agriculture Plant Pest Diagnostics Center. Special thanks to the Phoebe Apperson Hearst Museum staff, especially Joan Knudsen, Natasha Johnson, Ira Jacknis, and Thusa Chu for approving the project, helping to locate catalogue cards, and lending us seed samples from their collections. -

THE JEPSON GLOBE a Newsletter from the Friends of the Jepson Herbarium
THE JEPSON GLOBE A Newsletter from the Friends of The Jepson Herbarium VOLUME 29 NUMBER 1, Spring 2019 Curator’s column: Don Kyhos’s Upcoming changes in the Con- legacy in California botany sortium of California Herbaria By Bruce G. Baldwin By Jason Alexander In early April, my Ph.D. advisor, In January, the Northern California Donald W. Kyhos (UC Davis) turns 90, Botanists Association hosted their 9th fittingly during one of the California Botanical Symposium in Chico, Cali- desert’s most spectacular blooms in fornia. The Consortium of California recent years. Don’s many contributions Herbaria (CCH) was invited to present to desert botany and plant evolution on upcoming changes. The CCH be- in general are well worth celebrating gan as a data aggregator for California here for their critical importance to our vascular plant specimen data and that understanding of the California flora. remains its primary purpose to date. Those old enough to have used Munz’s From 2003 until 2017, the CCH grew A California Flora may recall seeing in size to over 2.2 million specimen re- the abundant references to Raven and cords from 36 institutions. Responding Kyhos’s chromosome numbers, which to requests from participants to display reflect a partnership between Don and specimen data from all groups of plants Peter Raven that yielded a tremendous Rudi Schmid at Antelope Valley Califor- and fungi, from all locations (including body of cytogenetic information about nia Poppy Reserve on 7 April 2003. Photo those outside California), we have de- our native plants. Don’s talents as a by Ray Cranfill.