The Use of Computational Methods for the Development of Molecularly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ford Talks on Preside.Ntial Disability Paper Deems Bv MATIHEW Coleman Paired
Cinematic SOcieties "CaVER·s:THE cAMPus un: THE MAGNaUAs." THuRsDAY, NOVEMBER 16,1995 Rat invaSion worries students, officials say it's not unusual '·' ' ·BY DANIEL JoHNsoN proceeded to lunge at Geppert. the east entrance to Reynolda Hall, outside of them before they capture us," Charlton said. length of eight inches, excluding the tail, and Cormueumm RFroRmR "l was coming home when a rat jumped at the Benson University Center and in the bu~hes He said he is particularly concerned about the weigh approximately one pound. me. So I just ran," Geppert said. He said he that surround Kitchin and Davis houses. large number of rats that he has seen between ''The students may be noticing a population Freshman Andy Oeppeit returned to his particularly feared that the rat would bite one 'Junior P.J. Charlton said-he has noticed an Davis and Benson. _ · bulge but it is not an abnormal occurrence," room in Kitchin House after a long njght of of his Birkenstock-exposed toes. increase in the amount of rats on campus The presence of rats on campus is not a new Weigl said. studying. Along his route from_ the library Geppert's experience is not unique." Over since h~ came t_o the university two years ago. problem. "There have always been rats on A surge in reproductio~ rates, more access back to Kitchin, Geppert encountered on~ of the past month, many students have noticed Charltonsaidhefeelsthatiftheratpopulation campus," said Peter Weigl, a professor of to food and shelter or movement to the cam the university rulers of the night: a-rat. -

Two-Dimensional Photonic Crystal Based Sensor for Pressure Sensing
PHOTONIC SENSORS / Vol. 4, No. 3, 2014: 248–253 Two-Dimensional Photonic Crystal Based Sensor for Pressure Sensing * Krishnan VIJAYA SHANTHI and Savarimuthu ROBINSON PG Scholar, Communication Systems, Mount Zion College of Engineering and Technology, Pudukkottai-622507, Tamil Nadu, India *Corresponding author: Krishnan VIJAYA SHANTHI Email: [email protected] Abstract: In this paper, a two-dimensional photonic crystal (2DPC) based pressure sensor is proposed and designed, and the sensing characteristics such as the sensitivity and dynamic range are analyzed over the range of pressure from 0 GPa to 7 GPa. The sensor is based on 2DPC with the square array of silicon rods surrounded by air. The sensor consists of two photonic crystal quasi waveguides and L3 defect. The L3 defect is placed in between two waveguides and is formed by modifying the radius of three Si rods. It is noticed that through simulation, the resonant wavelength of the sensor is shifted linearly towards the higher wavelength region while increasing the applied pressure level. The achieved sensitivity and dynamic range of the sensor is 2 nm/GPa and 7 Gpa, respectively. Keywords: Photonic crystal, waveguide, photonic band gap, optical sensor, FDTD method pressure sensor Citation: Krishnan VIJAYA SHANTHI and Savarimuthu ROBINSON, “Two-Dimensional Photonic Crystal Based Sensor for Pressure Sensing,” Photonic Sensors, 2014, 4(3): 248–253. one-dimensional photonic crystal (1DPCs), 1. Introduction two-dimensional photonic crystals (2DPCs), and In 1987, Eli Yablonovitch and Sajeev John three-dimensional photonic crystals (3DPCs). The published their research work on photonic crystals, sensor, based on 2DPCs, is receiving increasing predicting the existence of the photonic band gap as attention from the scientific community because it well as the potential for inhibiting spontaneous has relatively simple structure, small size, better emission and localizing light within defects in a confinement of light, accurate band gap calculation, and periodic lattice of appropriate dimensions [1]. -

KARLSSON (9781848443280) PRINT.Indd
Index accessibility, job vacancies 194, Augar, P. 288 197–207 Australia Acemoglu, D. 11, 13, 213 economic geography 161–2 Acha, V. 140 employment by industry sector Acs, Z.J. 52, 98, 139, 214 163–5, 185–6 Adams, P.D. 179 functional specialization 181–2 Adelaide 173, 178 growth determinants and forecasts Africa 307–8 178–81 agglomeration human capital 162, 171–2, 174, 175, economies 4, 51–2, 54–5, 282, 283, 176–7 321 income distribution, regional 177 human capital 2–7, 10 industry structure 162–70, 181–2 innovation in 65–6, 69–72 population and employment growth localized benefi ts 4 170–71 pecuniary externalities 70 rural unemployment 177 positive externalities 7–8 socioeconomic divides 162–3 Aghion, P. 11, 327 space economy 170, 189 agriculture, 105, 161, 162, 163, 164, technology industry 177–8 165, 166, 167, 174, 178, 179, 181, Australian Capital Territory 163, 171, 185, 328, 351, 356, 365 172 air travel 39 Australian Remoteness Classes Akerlof, G. 57 framework 164 Alfaro, L. 138, 220 Autant-Bernard, C. 15, 63, 76, 79, 81, Almeida, P. 73, 81, 82 89, 91 Altenburg, T. 138 Avci, G. 109 Amersfoort, van H. 109 Aw, B.Y. 321 Amin, A. 291, 294 Axtell, R. 143 Anastassova, L. 12 Andersson, Å.E. 8, 28, 29, 30, 31–2, babbling equilibrium 249 34, 41, 44, 46, 47, 49, 53, 301, 305 Balconi, M. 82 Andersson, M. 19, 194 Baldwin, R.E. 301 Anderstig, C. 42 Banerjee, A. 260 Angrist, J. 12, 13 banking sector 288, 295 Anselin, L. -

Babies' First Forenames: Births Registered in Scotland in 2009
Babies' first forenames: births registered in Scotland in 2009 Information about the basis of the list can be found via the 'Babies' First Names' page on the National Records of Scotland website. Boys Girls Position Name Number of babies Position Name Number of babies 1 Jack 706 1 Sophie 739 2 Lewis 656 2 Ava 486 3 James 472 3 Olivia 473 4 Logan 448 4 Emily 453 5 Liam 446 5= Chloe 433 6 Daniel 440 5= Lucy 433 7 Aaron 413 7 Katie 381 8 Ryan 393 8 Emma 376 9 Cameron 382 9 Erin 365 10 Callum 370 10 Amy 356 11 Alexander 349 11 Isla 354 12 Jamie 340 12 Ellie 337 13 Finlay 336 13 Jessica 313 14 Aiden 328 14 Hannah 301 15= Kyle 325 15 Lily 286 15= Lucas 325 16 Grace 280 17 Dylan 317 17 Eva 271 18 Matthew 300 18 Brooke 258 19 Adam 294 19 Holly 252 20 Nathan 292 20 Leah 240 21 Thomas 288 21 Mia 236 22 Ethan 275 22 Megan 231 23 Charlie 265 23 Millie 226 24 Oliver 253 24 Freya 212 25 Connor 244 25 Niamh 211 26 Max 240 26= Anna 210 27 Ben 237 26= Ruby 210 28 Joshua 230 28 Aimee 198 29 Jayden 229 29 Charlotte 195 30 Harry 226 30 Eilidh 193 31 William 223 31 Sarah 189 32 Michael 220 32 Abbie 180 33 Owen 219 33 Rebecca 179 34 Andrew 217 34 Lauren 170 35 Alfie 194 35 Zoe 164 36 Jay 193 36 Skye 163 37 David 192 37 Kayla 149 38 Joseph 187 38= Abigail 145 39 Samuel 186 38= Evie 145 40 Rhys 182 40 Molly 144 41 Ross 181 41 Summer 142 42 Tyler 174 42 Ella 140 43 John 173 43 Amelia 136 44 Rory 170 44 Amber 131 45 Kai 169 45 Rachel 126 46= Luke 167 46 Daisy 123 46= Scott 167 47 Eve 121 48 Sam 163 48 Caitlin 117 49 Archie 152 49 Cara 111 50 Euan 150 50 Zara 110 -

Four Sub-Types of Depression: a Self-Treatment Model
ARTICLE IN PRESS Medical Hypotheses xxx (2008) xxx–xxx Contents lists available at ScienceDirect Medical Hypotheses journal homepage: www.elsevier.com/locate/mehy Editorial A model for self-treatment of four sub-types of symptomatic ‘depression’ using non-prescription agents: Neuroticism (anxiety and emotional instability); malaise (fatigue and painful symptoms); demotivation (anhedonia) and seasonal affective disorder ‘SAD’ summary This article will present a model for how ‘depression’ (i.e. depressive symptoms) can be divided into four self-diagnosed sub-types or causes which might then be self-treated using agents available without prescription. (Another, much rarer, cause of depressed symptoms is the classical illness of ‘melancho- lia’, which when severe cannot be self-treated and typically requires hospitalization.) A self-management option and alternative is now needed due to the an inappropriate emphasis of modern psychiatry on treatment of imprecise syndromal ‘disorders’ which may entail treating ‘depression’ at the cost of making the patient feel and function worse. By contrast, the basic theoretical stance of self-management is that depressed mood should be seen as a result of unpleasant symptoms – and it is the symptoms that require treatment, not the mood itself. Furthermore, drugs (or other interventions) need to be clas- sified in terms of their potential therapeutic effects on these symptoms that may cause depressed mood. The four common causes of depressed mood considered here are the personality trait of Neuroticism; the state of malaise (fatigue, aching etc) which accompanies an illness with an activated immune system; demotivation due to lack of positive emotions (anhedonia); and the syndrome of seasonal affective disorder (SAD). -

Photonic Crystal Sensors Based on Porous Silicon
Sensors 2013, 13, 4694-4713; doi:10.3390/s130404694 OPEN ACCESS sensors ISSN 1424-8220 www.mdpi.com/journal/sensors Review Photonic Crystal Sensors Based on Porous Silicon Claudia Pacholski Department of New Materials and Biosystems, Max Planck Institute for Intelligent Systems, Heisenbergstr. 3, Stuttgart 70569, Germany; E-Mail: [email protected]; Tel.: +49-711-689-3620; Fax: +491-711-689-3612 Received: 8 February 2013; in revised form: 26 March 2013 / Accepted: 3 April 2013 / Published: 9 April 2013 Abstract: Porous silicon has been established as an excellent sensing platform for the optical detection of hazardous chemicals and biomolecular interactions such as DNA hybridization, antigen/antibody binding, and enzymatic reactions. Its porous nature provides a high surface area within a small volume, which can be easily controlled by changing the pore sizes. As the porosity and consequently the refractive index of an etched porous silicon layer depends on the electrochemial etching conditions photonic crystals composed of multilayered porous silicon films with well-resolved and narrow optical reflectivity features can easily be obtained. The prominent optical response of the photonic crystal decreases the detection limit and therefore increases the sensitivity of porous silicon sensors in comparison to sensors utilizing Fabry-Pérot based optical transduction. Development of porous silicon photonic crystal sensors which allow for the detection of analytes by the naked eye using a simple color change or the fabrication of stacked porous silicon photonic crystals showing two distinct optical features which can be utilized for the discrimination of analytes emphasize its high application potential. Keywords: photonic crystal; porous silicon; sensor 1. -

Unclaimed Capital Credits If Your Name Appears on This List, You May Be Eligible for a Refund
Unclaimed Capital Credits If your name appears on this list, you may be eligible for a refund. Complete a Capital Credits Request Form and return it to TCEC. 0'GRADY JAMES ABERNATHY RENETHA A ADAIR ANDY 2 C INVESTMENT ABETE LILLIE ADAIR CURTIS 2 GIRLS AND A COF FEE ABEYTA EUSEBIO ADAIR KATHLEEN SHOP ABEYTA SAM ADAIR LEANARD 21 DIESEL SERVICE ABLA RANDALL ADAIR SHAYLIN 4 RED CATTLE CO ABODE CONSTRUCTIO N ADAME FERNAND 5 C FARMS ABRAHA EDEN M ADAMS & FIELD 54 DINER ABRAHAM RAMPHIS ADAMS ALFRED 54 TOWING & RECOV ERY ABREM TERRY ADAMS ALVIN A & C FEED STORE ABSOLUTE ENDEAVORS INC ADAMS ALVIN L A & I SKYLINE ROO FING ACEVEDO ARNALDO ADAMS ASHLEY A & M CONSTRUCTIO N CO ACEVEDO DELFINA A ADAMS BAPTIST CHURCH A & S FARMS ACEVEDO LEONARD ADAMS CHARLES G A C CROSSLEY MOTE L ACKER EARL ADAMS DARREL A W H ON CO ACKER LLOYD ADAMS DAVID A WILD HAIR ACKER NELDA ADAMS DELBERT AARON KENNETH ACKERMAN AMIRAH ADAMS DIANE M ABADI TEAME ACKERS MELVIN D ADAMS DORIS ABARE JASMINE ACOSTA ARMANDO ADAMS DOYLE G ABARE KALYN ACOSTA CESAR ADAMS FERTILIZER ABAYNEH SOLOMON ACOSTA FERNANDO ADAMS GARY ABB RANDALL CORPO RATION ACOSTA GARCIA R ADAMS HALEY D ABBOTT CLYDE ACOSTA GILBERTO E ADAMS HEATHER ABBOTT CLYDE V ACOSTA JOE ADAMS JEROD ABBOTT FLOYD K ACOSTA JOSE A ADAMS JERRY ABBOTT MAUREAN ACOSTA MARIA ADAMS JILL M ABBOTT ROBERT ACOSTA VICTOR ADAMS JOHN ABBOTT SCOTTY ACTION REALTY ADAMS JOHNNY ACTON PAM ADAMS LINDA Current as of February 2021 Page 1 of 190 Unclaimed Capital Credits If your name appears on this list, you may be eligible for a refund. -

Teen Sensation Athing Mu
• ALL THE BEST IN RUNNING, JUMPING & THROWING • www.trackandfieldnews.com MAY 2021 The U.S. Outdoor Season Explodes Athing Mu Sets Collegiate 800 Record American Records For DeAnna Price & Keturah Orji T&FN Interview: Shalane Flanagan Special Focus: U.S. Women’s 5000 Scene Hayward Field Finally Makes Its Debut NCAA Formchart Faves: Teen LSU Men, USC Women Sensation Athing Mu Track & Field News The Bible Of The Sport Since 1948 AA WorldWorld Founded by Bert & Cordner Nelson E. GARRY HILL — Editor JANET VITU — Publisher EDITORIAL STAFF Sieg Lindstrom ................. Managing Editor Jeff Hollobaugh ................. Associate Editor BUSINESS STAFF Ed Fox ............................ Publisher Emeritus Wallace Dere ........................Office Manager Teresa Tam ..................................Art Director WORLD RANKINGS COMPILERS Jonathan Berenbom, Richard Hymans, Dave Johnson, Nejat Kök SENIOR EDITORS Bob Bowman (Walking), Roy Conrad (Special AwaitsAwaits You.You. Projects), Bob Hersh (Eastern), Mike Kennedy (HS Girls), Glen McMicken (Lists), Walt Murphy T&FN has operated popular sports tours since 1952 and has (Relays), Jim Rorick (Stats), Jack Shepard (HS Boys) taken more than 22,000 fans to 60 countries on five continents. U.S. CORRESPONDENTS Join us for one (or more) of these great upcoming trips. John Auka, Bob Bettwy, Bret Bloomquist, Tom Casacky, Gene Cherry, Keith Conning, Cheryl Davis, Elliott Denman, Peter Diamond, Charles Fleishman, John Gillespie, Rich Gonzalez, Ed Gordon, Ben Hall, Sean Hartnett, Mike Hubbard, ■ 2022 The U.S. Nationals/World Champion- ■ World Track2023 & Field Championships, Dave Hunter, Tom Jennings, Roger Jennings, Tom ship Trials. Dates and site to be determined, Budapest, Hungary. The 19th edition of the Jordan, Kim Koffman, Don Kopriva, Dan Lilot, but probably Eugene in late June. -

Most-Common-Surnames-Bmd-Registers-16.Pdf
Most Common Surnames Surnames occurring most often in Scotland's registers of Births, Marriages and Deaths Counting only the surname of the child for births, the surnames of BOTH PARTIES (for example both BRIDE and GROOM) for marriages, and the surname of the deceased for deaths Note: the surnames from these registers may not be representative of the surnames of the population of Scotland as a whole, as (a) they include the surnames of non-residents who were born / married / died here; (b) they exclude the surnames of residents who were born / married / died elsewhere; and (c) some age-groups have very low birth, marriage and death rates; others account for most births, marriages and deaths.ths Registration Year = 2016 Position Surname Number 1 SMITH 2056 2 BROWN 1435 3 WILSON 1354 4 CAMPBELL 1147 5 STEWART 1139 6 THOMSON 1127 7 ROBERTSON 1088 8 ANDERSON 1001 9 MACDONALD 808 10 TAYLOR 782 11 SCOTT 771 12 REID 755 13 MURRAY 754 14 CLARK 734 15 WATSON 642 16 ROSS 629 17 YOUNG 608 18 MITCHELL 601 19 WALKER 589 20= MORRISON 587 20= PATERSON 587 22 GRAHAM 569 23 HAMILTON 541 24 FRASER 529 25 MARTIN 528 26 GRAY 523 27 HENDERSON 522 28 KERR 521 29 MCDONALD 520 30 FERGUSON 513 31 MILLER 511 32 CAMERON 510 33= DAVIDSON 506 33= JOHNSTON 506 35 BELL 483 36 KELLY 478 37 DUNCAN 473 38 HUNTER 450 39 SIMPSON 438 40 MACLEOD 435 41 MACKENZIE 434 42 ALLAN 432 43 GRANT 429 44 WALLACE 401 45 BLACK 399 © Crown Copyright 2017 46 RUSSELL 394 47 JONES 392 48 MACKAY 372 49= MARSHALL 370 49= SUTHERLAND 370 51 WRIGHT 357 52 GIBSON 356 53 BURNS 353 54= KENNEDY 347 -

Silicon Photonic Crystal Microarrays for High Throughput Label-Free
Invited Paper Silicon photonic crystal microarrays for high throughput label-free detection of lung cancer cell line lysates with sensitivity and specificity Swapnajit Chakravartya, Wei-Cheng Laib, Yi Zoub, Robert M. Gemmillc, Ray T. Chen*a,b, aOmega Optics Inc., 10306 Sausalito Drive, Austin, TX, USA 78759; bDept. of Electrical and Computer Engineering, University of Texas, 10100 Burnet Road Bldg. 160, Austin, TX USA 78758; cMedical University of South Carolina, 96 Jonathan Lucas Street, Charleston, SC 29425; ABSTRACT Detection of biomolecules on microarrays based on label-free on-chip optical biosensors is very attractive since this format avoids complex chemistries caused by steric hindrance of labels. Application areas include the detection of cancers and allergens, and food-borne pathogens to name a few. We have demonstrated photonic crystal microcavity biosensors with high sensitivity down to 1pM concentrations (67pg/ml). High sensitivities were achieved by slow light engineering which reduced the radiation loss and increased the stored energy in the photonic crystal microcavity resonance mode. Resonances with high quality factor Q~26,760 in liquid ambient, coupled with larger optical mode volumes allowed enhanced interaction with the analyte biomolecules which resulted in sensitivities down to 10 cells per micro-liter to lung cancer cell lysates. The specificity of detection was ensured by multiplexed detections from multiple photonic crystal microcavities arrayed on the arms of a multimode interference power splitter. Specific binding interactions and control experiments were performed simultaneously at the same instant of time with the same 60 micro- liter sample volume. Specificity is further ensured by sandwich assay methods in the multiplexed experiment. -
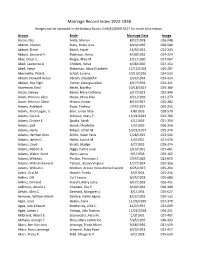
Marriage Record Index 1922-1938 Images Can Be Accessed in the Indiana Room
Marriage Record Index 1922-1938 Images can be accessed in the Indiana Room. Call (812)949-3527 for more information. Groom Bride Marriage Date Image Aaron, Elza Antle, Marion 8/12/1928 026-048 Abbott, Charles Ruby, Hallie June 8/19/1935 030-580 Abbott, Elmer Beach, Hazel 12/9/1922 022-243 Abbott, Leonard H. Robinson, Berta 4/30/1926 024-324 Abel, Oscar C. Ringle, Alice M. 1/11/1930 027-067 Abell, Lawrence A. Childers, Velva 4/28/1930 027-154 Abell, Steve Blakeman, Mary Elizabeth 12/12/1928 026-207 Abernathy, Pete B. Scholl, Lorena 10/15/1926 024-533 Abram, Howard Henry Abram, Elizabeth F. 3/24/1934 029-414 Absher, Roy Elgin Turner, Georgia Lillian 4/17/1926 024-311 Ackerman, Emil Becht, Martha 10/18/1927 025-380 Acton, Dewey Baker, Mary Cathrine 3/17/1923 022-340 Adam, Herman Glen Harpe, Mary Allia 4/11/1936 031-273 Adam, Herman Glenn Hinton, Esther 8/13/1927 025-282 Adams, Adelbert Pope, Thelma 7/14/1927 025-255 Adams, Ancil Logan, Jr. Eiler, Lillian Mae 4/8/1933 028-570 Adams, Cecil A. Johnson, Mary E. 12/21/1923 022-706 Adams, Crozier E. Sparks, Sarah 4/1/1936 031-250 Adams, Earl Snook, Charlotte 1/5/1935 030-250 Adams, Harry Meyer, Lillian M. 10/21/1927 025-376 Adams, Herman Glen Smith, Hazel Irene 2/28/1925 023-502 Adams, James O. Hallet, Louise M. 4/3/1931 027-476 Adams, Lloyd Kirsch, Madge 6/7/1932 028-274 Adams, Robert A. -
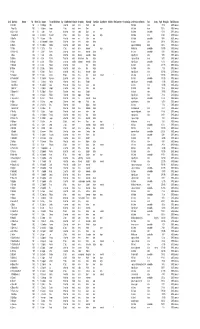
Surnames Beginning With
Card# Mine Name Operator Year Month Day Surname First and Middle Name Age Fatal/Nonfatal In/Outside Occupation Nationality Citizen/Alien Single/Married #Children Mine Experience Occupation Exp. Accident Cause or Remarks Fault County Page# Mining Dist. Film# Bituminous 33 Kerr No.8 1901 9 17 Cabbage Mike 42 nonfatal inside miner Polish alien fall of slate victim 73 3rd 3593 Bituminous 3 Fayett City 1902 1 4 Cabitcher Joseph 17 fatal inside miner Slavic single fall of slate others 100 1st 3593 Bituminous 146 Somers No.2 1901 10 1 Cabor Adam 66 nonfatal inside loader Slavic alien fall of slate unavoidable 67 1st 3593 Bituminous 5 Clarion No.27 1906 1 22 Cacallito Domenico 34 fatal inside miner Italian alien single fall of slate victim 213 4th 3593 Bituminous 30 Bear Run 1900 11 5 Cacara Mike 43 nonfatal inside mm Italian alien fall of slate unavoidable 55 8th 3593 Bituminous 12 Fowler 1906 2 26 Cadanyaretti Michael 37 nonfatal inside miner Italian citizen fall of roof victim 209 3rd 3593 Bituminous 146 Beaznell 1905 11 15 Caddock Charles 29 nonfatal inside laborer Slavic alien explosion of fire damp others 134 1st 3593 Bituminous 75 Trotter 1905 11 11 Caffery Peter 19 fatal inside driver Amercian single killed by cars unavoidable 166 10th 3593 Bituminous 40 Webster No.3 1901 8 2 Cahill Andrew 24 nonfatal inside miner Amercian citizen fall of coal unavoidable 82 6th 3593 Bituminous 2 Hazel 1904 1 8 Cain John L 27 nonfatal inside foreman Amercian citizen injured by cars victim 90 7th 3593 Bituminous 100 Dickson 1904 10 4 Cain Patrick 75 nonfatal