10 August 2021 Aperto
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Publication of an Application for Approval of a Minor Amendment in Accordance with the Second Subparagraph of Article 53(2)
30.8.2016 EN Official Journal of the European Union C 315/3 OTHER ACTS EUROPEAN COMMISSION Publication of an application for approval of a minor amendment in accordance with the second subparagraph of Article 53(2) of Regulation (EU) No 1151/2012 of the European Parliament and of the Council (2016/C 315/03) The European Commission has approved this minor amendment in accordance with the third subparagraph of Article 6(2) of Commission Delegated Regulation (EU) No 664/2014 (1). APPLICATION FOR APPROVAL OF A MINOR AMENDMENT Application for approval of a minor amendment in accordance with the second subparagraph of Article 53(2) of Regulation (EU) No 1151/2012 of the European Parliament and of the Council (2) ‘BRA’ EU No: PDO-IT-02128 — 18.3.2016 PDO ( X ) PGI ( ) TSG ( ) 1. Applicant group and legitimate interest Consorzio di Tutela del Formaggio BRA DOP (association for the protection of ‘Bra’ PDO cheese) Via Silvio Pellico 10 10022 Carmagnola (TO) ITALIA Tel. +39 0110565985. Fax +39 0110565989. Email: [email protected] The Consorzio di Tutela del Formaggio BRA DOP is entitled to submit an amendment application pursuant to Article 13(1) of Ministry of Agricultural, Food and Forestry Policy Decree No 12511 of 14 October 2013. 2. Member State or Third Country Italy 3. Heading in the product specification affected by the amendment(s) — Product description — Proof of origin — Production method — Link — Labelling — Other [to be specified] 4. Type of amendment(s) — Amendment to the product specification of a registered PDO or PGI to be qualified as minor in accordance with the third subparagraph of Article 53(2) of Regulation (EU) No 1151/2012, that requires no amend ment to the published single document. -

Pag Chr Prova
Granda bike tour DE VICOFORTE Granda À MONDOVÌ 1 ET ROCCAFORTE DÉPART Au coeur des vallées de Mondovì, le ha- Vicoforte (512 m) meau caractéristique de Prea (838 m) et ARRIVÉE bike tour après 3 km de dure côte le Sanctuaire de Roccaforte (570 m) Qui est convaincu qu’une des façons les Sant’Anna, exemple de baroque français de plus valables pour connaître le dépar- 1763 (1098 m) méritent le détour. Du Sanc- LONGUEUR tement de Cuneo est de l’explorer en tuaire de Vicoforte, en parcourant à gauche km 34,000 bicyclette aura l’occasion d’apprécier un le périple de la Basilique, puis viale Marconi, on DÉNIVELÉ produit qui l’enthousiasmera pour le se dirige vers Fiamenga, puis à Mondovì le m 600 long de la “Via delle Cappelle” (route des cha- grand éventail d’offres disponibles. Sur ALTITUDE MAXIMUM la selle d’un vélo de route on peut jouir pelles). On suit la direction de Via dell’Annun- ziata sur la côte pour San Lorenzo, vers Mona- m 650 d’une perspective tout à fait différente. stero Vasco. La direction maintenant est DIFFICULTÉ Cette splendide partie du nord de l’Italie Montaldo, dans la Val Corsaglia et après case Moyenne-Difficile n’est pas plate: d’où les panoramas mer- Giacobba, 4 km de côte, à droite par Via La- veilleux et c’est pour cela que l’on re- rone Balun-Luva. On dépasse les Bertolini commande une préparation psychophy- Soprani, à gauche et après 1 km à droite. On sique adéquate. Deux sorties par semai- continue ensuite à gauche Via Unie en descen- ne avec quelques côtes seront certaine- te, on arrive ainsi Via Alma qui se parcourra en contournant Madonna della Neve. -
Presepiingranda.It
NATALE AVVISO2015 SACRO IL PRESEPE TRA ARTE, FEDE E TRADIZIONE Itinerari natalizi a Cuneo e Provincia INGRESSO LIBERO SCOPRI TUTTI I PRESEPI SU WWW.presepiingranda.it a partecipazione di ognuno al servizio di tutti è l’intento principale di Presepi in Granda un progetto Lnato nel Natale 2005 per promuovere e diffondere il messaggio, la passione e la tradizione del presepe a Cuneo e Provincia. Presepi in Granda è un’iniziativa libera e aperta a tutti, contattaci per informarti: [email protected] · +39 328 2637000. FARIGLIANO ALBA 22/11 - 6/01 SAN PIETRO DEL GALLO 13/12 - 6/01 GARESSIO 25/12 - 31/01 BANDITO DI BRA 23/12 - 6/01 SAVIGLIANO 25/12 - 6/01 ISASCA 23/12 - 6/01 BORGO S. DALMAZZO 24/12 - 31/01 SCARNAFIGI 13/12 - 27/12 MELLE 8/12 - 10/01 BOVES 24/12 - 31/01 TARANTASCA 13/12 - 6/01 MONDOVÌ 20/12 - 31/01 BRA 13/12 - 6/01 TETTI DI DRONERO 13/12 - 6/01 PIANFEI 25/12 - 17/01 BUSCA 20/12 - 10/01 VALDIERI 19/12 - 31/01 RACCONIGI 12/12 - 10/01 CASTELLAR di BOVES 6/12 - 10/01 VERDUNO 8/12 - 24/01 RICCA S. ROCCO CHERASCA 8/12 - 17/01 CAVALLERMAGGIORE 26/12 - 10/01 VERGNE 8/12 - 10/01 RUFFIA 22/12 - 6/01 CUNEO SANTA CHIARA 5/12 - 17/01 VERNANTE 20/12 - 6/01 SAMPEYRE 24/12 - 31/01 CUNEO SAN PAOLO 24/12 - 24/01 VILLANOVA SOLARO 20/12 - 31/01 SAN BERNARDO DI CERVASCA 13/12 - 17/01 DIANO D’ALBA 26/12 - 17/01 CARMAGNOLA 13/12 - 31/01 SAN GIACOMO DI BOVES 25/12 - 6/01 ENTRACQUE 20/12 - 10/01 VIGONE 24/12 - 6/01 SANFRÈ 6/12 - 10/01 25/12 - 10/01 Si declina ogni responsabilità da eventuali variazioni di aperture e orari dei singoli presepi. -

Cinque Passi Tra Basso Piemonte E Francia Sampeyre È Un Ridente Paesino Della Valle Varaita in Provincia Di Cuneo Scelto Da
Cinque passi tra Basso Piemonte e Francia Sampeyre è un ridente paesino della Valle Varaita in provincia di Cuneo scelto da noi come punto di partenza per accompagnarvi in questo tour in moto tra gli spettacolari passi delle Alpi: Colle di Sampeyre, Colle Fauniera, Col de Larche, Col de Vars e Colle Agnello al confine fra Basso Piemonte e la Francia. Questa è una zona non molto frequentata da motociclisti che preferiscono la confusione dei valichi dell'alta Lombardia, Veneto e Trentino, straordinari per i panorami mozzafiato che sanno regalare, ma vi consigliamo di seguire questi percorsi attraverso le Alpi Marittime e vi renderete conto di avere trovato un altro paradiso per motociclisti. Da Sampeyre seguite le indicazioni per Sodani, Goria, Elva e Colle di Sampeyre per poi salire, superando 7 tornanti, ai 2.284 metri di quota del colle dove potrete ammirare una veduta grandiosa sul Monviso e su tutte le Alpi circostanti. In un prossimo percorso parleremo dell'adiacente "Orrido di Elva", una stretta e sinuosa strada scavata nella roccia che lascia sempre affascinati tutti quelli che l'hanno percorsa. Scendiamo poi in direzione di Ponte di Marmora per poi iniziare la salita verso il Colle dei Morti, chiamato anche Colle Fauniera, che ci porterà a raggiungere i 2.481 m. di altezza seguendo una strada stretta e non molto in buone condizioni, ma la bellezza dell'ambiente circostante è tale da farci dimenticare tutto. La discesa dal Colle dei Morti ci porta diritto a Demonte da cui deviamo a destra sulla statale 21 seguendo le indicazioni per Vinadio, Pontebernardo e, appena superata Argentera, ecco davanti a noi i tornanti del Colle della Maddalena o Col de Larche come lo chiamano i francesi. -

Senza Titolo
Criteri stima di gravità e fattibilità Le stelle * indicano da 1= minimo a 6= massimo il grado di fattibilità e gravità (degrado ambientale) dei siti censiti. La valutazione della fattibilità (indice che rappresenta la possibilità di bonificare il sito) è stata effettuata seguendo i seguenti criteri: 1) Possibilità e/o impegno da parte degli enti locali 2) Raggiungibilità da parte di automezzi (presenza di strade sterrate) per effettuare lavori di bonifica 3) Possibile sostegno percepito da interviste con abitanti del luogo 4) Possibilità di riconvertire e/o riutilizzare le strutture edificate Per quanto concerne la valutazione della gravità: 1) Grado di abbandono 2) Pericoli per la sicurezza degli escursionisti e dei cittadini 3) Impatto visivo 4) Presenza di materiali inquinanti 5) Possibilità di progetti che contemplino la riqualifica anziché la bonifica 6) Siti in parchi naturali o in zone di pregio naturalistico. Pro Natura - Censimento impianti abbandonati Piemonte – Mountain Wilderness Scheda 1 Vandalino Torre Pellice (TO) Località: Monte Vandalino –Sea di Torre (Valle Pellice) Accesso: da Torre Pellice o dalla Valle di Angrogna Comune: Torre Pellice Tipologia impianti: seggiovia(tipo gabbiotti, smantellata, restano solo i tralicci) e skilift Strutture presenti: Stazioni di partenza e di arrivo, tralicci dell’impianto di arroccamento, gabbiotto di partenza di un piccolo skilift. La stazione di partenza è ora in fase di ristrutturazione, ad uso residenziale. Contesto, altitudine ed esposizione: la stazione di partenza è nei pressi della Provinciale; l’arrivo e il piccolo skilift sono situati sulla dorsale tra la valle principale e la valle di Angrogna ad una quota di circa 1200 metri. -

Scarica L'appendice Con L'elenco Completo Degli Edifici Dismessi
Appendice II del Quaderno 37 della Collana della Fondazione CRC Rigenerare spazi dismessi Nuove prospettive per la comunità Elenco completo dei beni dismessi rilevati in provincia di Cuneo attraverso le fonti e grazie alle segnalazioni degli enti territoriali (aggiornato al 18 giugno 2018) Nota metodologica A cura di Fondazione Fitzcarraldo Si rende disponibile l’elenco dei 449 beni dismessi in provincia di Cu- neo rilevati attraverso le fonti disponibili e grazie alle segnalazioni dirette degli enti territoriali nell’ambito della mappatura realizzata per il Quaderno 37 Rigenerare spazi dismessi. Nuove prospettive per la comunità, pro- mosso dalla Fondazione CRC e realizzato da Fondazione Fitzcarraldo. Di seguito si specificano alcune considerazioni relative alla metodolo- gia adottata e alcune indicazioni utili alla consultazione dell’elenco. La scelta effettuata per disporre di una prima ricognizione di parte del patrimonio dismesso cuneese si è concretata, da un lato, in una ricerca e una lettura dei dati estrapolati dalle poche fonti disponibili e, dall’altro, nella raccolta di segnalazioni dirette da parte degli enti territoriali. Una mappatura efficace e puntuale in grado di fotografare dal punto di vista quantitativo e localizzativo tutte le opportunità e gli edifici in stato di abbandono e di sottoutilizzo in un territorio così vasto come la provincia di Cuneo non può prescindere da sopralluoghi mirati in tutti i 250 comuni del territorio. Per questo è più realistico definire questa parte della ricerca come una ricognizione “a maglie larghe” di un immenso patrimonio di- smesso che comprende una moltitudine di proprietari differenti. Le diverse fonti oggetto della presente ricognizione individuano beni le cui condizioni effettive non sono direttamente verificate, non essendo stati effettuati i sopralluoghi. -
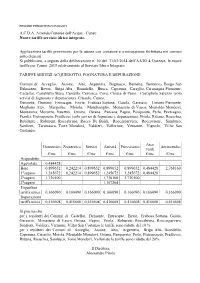
Aa Aa A.C.D.A
REGIONE PIEMONTE BU5 05/02/2015 A.C.D.A. Azienda Cuneese dell'Acqua - Cuneo Nuove tariffe servizio idrico integrato. Applicazione tariffe provvisorie per le utenze con contatore e a misurazione forfettaria nei comuni sotto elencati. Si pubblicano, a seguito della deliberazione n. 10 del 31/03/2014 dell’AATO 4 Cuneese, le nuove tariffe per l’anno 2015 relativamente al Servizio Idrico Integrato TARIFFE SERVIZI ACQUEDOTTO, FOGNATURA E DEPURAZIONE Comuni di: Acceglio, Aisone, Alto, Argentera, Bagnasco, Beinette, Bernezzo, Borgo San Dalmazzo, Boves, Briga Alta, Brondello, Busca, Caprauna, Caraglio, Caramagna Piemonte, Castellar, Castelletto Stura, Centallo, Cervasca, Ceva, Chiusa di Pesio, Costigliole Saluzzo (solo servizi di fognatura e depurazione), Crissolo, Cuneo, Demonte, Dronero, Entracque, Envie, Frabosa Sottana, Gaiola, Garessio, Limone Piemonte, Magliano Alpi, Margarita, Moiola, Mombasiglio, Monastero di Vasco, Montaldo Mondovì, Montanera, Morozzo, Nucetto, Ormea, Ostana, Paesana, Pagno, Pamparato, Perlo, Peveragno, Pianfei, Pietraporzio, Pradleves (solo servizi di fognatura e depurazione), Priola, Rittana, Roaschia, Robilante , Roburent, Roccabruna, Rocca De Baldi, Roccasparvera, Roccavione, Sambuco, Sanfront, Tarantasca, Torre Mondovì, Valdieri, Valloriate, Vernante, Vignolo, Villar San Costanzo. Aree Domestico Zootecnico Servizi Attività Provvisorio Antincendio verdi €/mc €/mc €/mc €/mc €/mc €/mc €/mc Acquedotto Agevolata 0,484428 Base 0,899652 0,242214 0,899652 0,899652 0,899652 0,484428 2,768160 1°supero 1,245672 0,242214 0,899652 -
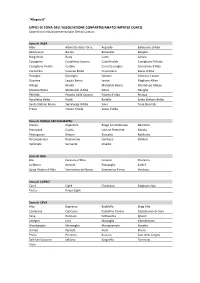
Zone Del Sistema Confartigianato Cuneo -> Comuni
“Allegato B” UFFICI DI ZONA DELL’ASSOCIAZIONE CONFARTIGIANATO IMPRESE CUNEO Zone e loro limitazione territoriale. Elenco Comuni. Zona di ALBA Alba Albaretto della Torre Arguello Baldissero d’Alba Barbaresco Barolo Benevello Bergolo Borgomale Bosia Camo Canale Castagnito Castelletto Uzzone Castellinaldo Castiglione Falletto Castiglione Tinella Castino Cerretto Langhe Corneliano d’Alba Cortemilia Cossano Belbo Cravanzana Diano d’Alba Feisoglio Gorzegno Govone Grinzane Cavour Guarene Lequio Berria Levice Magliano Alfieri Mango Montà Montaldo Roero Montelupo Albese Monteu Roero Monticello d’Alba Neive Neviglie Perletto Pezzolo Valle Uzzone Piobesi d’Alba Priocca Rocchetta Belbo Roddi Rodello Santo Stefano Belbo Santo Stefano Roero Serralunga d’Alba Sinio Tone Bormida Treiso Trezzo Tinella Vezza d’Alba Zona di BORGO SAN DALMAZZO Aisone Argentera Borgo San Dalmazzo Demonte Entracque Gaiola Limone Piemonte Moiola Pietraporzio Rittana Roaschia Robilante Roccasparvera Roccavione Sambuco Valdieri Valloriate Vernante Vinadio Zona di BRA Bra Ceresole d’Alba Cervere Cherasco La Morra Narzole Pocapaglia Sanfrè Santa Vittoria d’Alba Sommariva del Bosco Sommariva Perno Verduno Zona di CARRÙ Carrù Cigliè Clavesana Magliano Alpi Piozzo Rocca Cigliè Zona di CEVA Alto Bagnasco Battifollo Briga Alta Camerana Caprauna Castellino Tanaro Castelnuovo di Ceva Ceva Garessio Gottasecca Igliano Lesegno Lisio Marsaglia Mombarcaro Mombasiglio Monesiglio Montezemolo Nucetto Ormea Paroldo Perlo Priero Priola Prunetto Roascio Sale delle Langhe Sale San Giovanni Saliceto -

Maira Stradale Botte
Pontechianale Serro Maddalena Saluzzo Castello Gambasca Martiniana Maurin Borga Pagno Caldane Bianchi S.Anna Villar Dragoniere Casteldelfino Becetto Brodello Ve 1882 Belllino Sampeyre Masueira Torrete M.Maniglia Rore Isasca www.locandamistral.com •3177 )(Colle Bicocca Frassino 2637 Pratolungo Brossasco Brec de )( Torr Colle Maurin S.Annna ente Varaita Chambeyron Melle Piasco •3411 )( Colle di Sampeyre S.Maurizio S.Antonio Chiosso Venasca Elva Serre Valmala Cerreto Chesta Rossana Rocca Cucchiales Provenzale Eremo Bersia Lemma S.Michele Prazzo Stroppo Busca M.Sautron Chiappera Bedale Camoglieres •3166 Ussolo Ponte Maira Torrente Maira Macra Lottulo Prazzo ra Ponte Marmora ai Acceglio Bassura San Damiano Macra M M.Soubeyran te S.Giuseppe Locanda n 2701 Celle di Macra Roccabruna re )( MISTRAL Maddalena Albareto Villar San r Pas.della Cavalla o Costanzo T Morra Combamala DRONERO Marmora Cartignano Canosio Chialvetta Reinero M.Oronaye Paglieres Tetti •3100 S.Margherita Pratorotondo Pratavecchia Villata Viviere Tolosano Allardo 1997 Preit Colle della Maddalena Montemale P.della Gardetta )( Pas.Peroni )( S.Lorenzo Pradleves S.Giorgio )( Grangie a Castelmagno n M.VentasusoGite MTB Argenterdalla a Col Osero Cavaliggi Caraglio ra •2701 na e G Rua Bernardo Locanda Mistral S.Magno Campomolino e Gra nt Colle Colle Torrent rre 01 Xc Experience )( Valgrana To San Bernardo Margherita d’Eschiele Chiappi 02 bassa valle )( Chiotti Monterosso 03 Dragon Bike Bersezio Frise Grana 04 Colli Birrone, Melle, Ciabra 05 Strada dei Cannoni Figliere S.Anna 06 Rocca -

Deliberazione Della Giunta Regionale 27 Maggio 2019, N. 35-9084 LR 18
REGIONE PIEMONTE BU26 27/06/2019 Deliberazione della Giunta Regionale 27 maggio 2019, n. 35-9084 L.R. 18/2017 - approvazione dello schema di Accordo di programma tra la Regione Piemonte e il Comune di Frabosa Soprana per la realizzazione dell'intervento di "Potenziamento dell'impianto di innevamento programmato della stazione sciistica di Frabosa Soprana". Spesa regionale Euro 800.000,00. A relazione dell'Assessore Parigi: Premesso che: con l’art. 13 della legge regionale 22 novembre 2017, n. 18 “Assestamento di bilancio di previsione finanziario 2017/2019 e disposizioni finanziarie”, è stato istituito presso Finpiemonte S.p.A. un fondo di € 24.500.000,00 destinato al sostegno di investimenti relativi a progetti di sviluppo turistico dei territori montani tramite: - Accordi di programma, da stipularsi tra la Regione Piemonte e gli Enti locali ai sensi dell’art. 34 del D.Lgs 267/00 e s.m.i. e della D.G.R. n. 27 – 23223 del 24/11/1997 modificata con D.G.R. n. 1 – 7327 del 03.08.2018; - concessione di garanzie su finanziamenti da parte di imprese private; la Giunta regionale, con deliberazione n. 48 – 6154 del 15 dicembre 2017, come da ultimo modificata con D.G.R. 22 febbraio 2019, n. 26-8452, ha approvato i criteri per la sottoscrizione di Accordi di programma volti al sostegno di interventi per la creazione ed il potenziamento del turismo montano invernale ed estivo riconoscendo prioritari gli interventi relativi ai seguenti ambiti: - investimenti inerenti l’innevamento programmato; - investimenti inerenti gli impianti di risalita; - altri investimenti inerenti il potenziamento e la rivitalizzazione del turismo montano sia invernale sia estivo. -

Second Report Submitted by Italy Pursuant to Article 25, Paragraph 1 of the Framework Convention for the Protection of National Minorities
Strasbourg, 14 May 2004 ACFC/SR/II(2004)006 SECOND REPORT SUBMITTED BY ITALY PURSUANT TO ARTICLE 25, PARAGRAPH 1 OF THE FRAMEWORK CONVENTION FOR THE PROTECTION OF NATIONAL MINORITIES (received on 14 May 2004) MINISTRY OF THE INTERIOR DEPARTMENT FOR CIVIL LIBERTIES AND IMMIGRATION CENTRAL DIRECTORATE FOR CIVIL RIGHTS, CITIZENSHIP AND MINORITIES HISTORICAL AND NEW MINORITIES UNIT FRAMEWORK CONVENTION FOR THE PROTECTION OF NATIONAL MINORITIES II IMPLEMENTATION REPORT - Rome, February 2004 – 2 Table of contents Foreword p.4 Introduction – Part I p.6 Sections referring to the specific requests p.8 - Part II p.9 - Questionnaire - Part III p.10 Projects originating from Law No. 482/99 p.12 Monitoring p.14 Appropriately identified territorial areas p.16 List of conferences and seminars p.18 The communities of Roma, Sinti and Travellers p.20 Publications and promotional activities p.28 European Charter for Regional or Minority Languages p.30 Regional laws p.32 Initiatives in the education sector p.34 Law No. 38/2001 on the Slovenian minority p.40 Judicial procedures and minorities p.42 Database p.44 Appendix I p.49 - Appropriately identified territorial areas p.49 3 FOREWORD 4 Foreword Data and information set out in this second Report testify to the considerable effort made by Italy as regards the protection of minorities. The text is supplemented with fuller and greater details in the Appendix. The Report has been prepared by the Ministry of the Interior – Department for Civil Liberties and Immigration - Central Directorate for Civil Rights, Citizenship and Minorities – Historical and new minorities Unit When the Report was drawn up it was also considered appropriate to seek the opinion of CONFEMILI (National Federative Committee of Linguistic Minorities in Italy). -

Tentacular Thinking: Anthropocene, Warming and Acidification of the Oceans from Capitalocene, Chthulucene,” in Donna J
We are all lichens. – Scott Gilbert, “We Are All Lichens Now”1 Think we must. We must think. – Stengers and Despret, Women Who Make 01/17 a Fuss2 What happens when human exceptionalism and bounded individualism, those old saws of Western philosophy and political economics, become unthinkable in the best sciences, whether natural or social? Seriously unthinkable: not available to think with. Biological sciences Donna Haraway have been especially potent in fermenting notions about all the mortal inhabitants of the Earth since the imperializing eighteenth century. Tentacular Homo sapiens – the Human as species, the Anthropos as the human species,Modern Man – Thinking: was a chief product of these knowledge practices. What happens when the best biologies Anthropocene, of the twenty-first century cannot do their job with bounded individuals plus contexts, when organisms plus environments, or genes plus Capitalocene, whatever they need, no longer sustain the overflowing richness of biological knowledges, if Chthulucene they ever did? What happens when organisms plus environments can hardly be remembered for the same reasons that even Western-indebted people can no longer figure themselves as individuals and societies of individuals in e n human-only histories? Surely such a e c transformative time on Earth must not be named u l u the Anthropocene! h y t a h ÊÊÊÊÊÊÊÊÊÊWith all the unfaithful offspring of the sky C w , a r e gods, with my littermates who find a rich wallow a n e H c in multispecies muddles, I want to make a a o n l a n critical and joyful fuss about these matters.