Molecular Phylogenetics of the Buteonine Birds of Prey (Accipitridae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Buteogallus Coronatus) in Argentina
SHORT COMMUNICATIONS J. Raptor Res. 54(2):166–171 Ó 2020 The Raptor Research Foundation, Inc. ELECTROCUTION ON POWER LINES IS AN IMPORTANT THREAT FOR THE ENDANGERED CHACO EAGLE (BUTEOGALLUS CORONATUS) IN ARGENTINA 1 JOSE´ H. SARASOLA Centro para el Estudio y Conservacio´n de Aves Rapaces en Argentina (CECARA). Universidad Nacional de La Pampa, Avda Uruguay 151, 6300 Santa Rosa, Argentina and Instituto de las Ciencias Ambientales y de la Tierra de La Pampa (INCITAP-CONICET), Avda Uruguay 151, 6300 Santa Rosa, La Pampa, Argentina MAXIMILIANO A. GALMES Centro para el Estudio y Conservacio´n de Aves Rapaces en Argentina (CECARA), Universidad Nacional de La Pampa, Avda Uruguay 151, 6300 Santa Rosa, Argentina and The Peregrine Fund, Boise, ID 83709 USA BRYAN D. WATTS Center for Conservation Biology, College of William & Mary, Williamsburg, VA 23187 USA and Virginia Commonwealth University, Williamsburg, VA 23284 USA ABSTRACT.—Electrocution is a widespread conservation problem for birds of prey that has received little attention in the Neotropics. Here we present electrocution records involving the endangered Chaco Eagle (Buteogallus coronatus) in central Argentina, and we provide information on the power pole structural characteristics associated with electrocutions. Nine Chaco Eagles were recorded electrocuted during the period 2012–2019 over an area of 9000 km2. Chaco Eagles were found electrocuted in association with five types of power poles, but more than half the electrocutions (55%) were on poles made of steel-reinforced concrete and with jumper wires above the crossarms. With the addition of four previous electrocution reports in this region during the same time period, the annual rate of Chaco Eagle electrocutions was similar to the rate of mortality by other human-related factors such as direct persecution. -

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

A Multi-Gene Phylogeny of Aquiline Eagles (Aves: Accipitriformes) Reveals Extensive Paraphyly at the Genus Level
Available online at www.sciencedirect.com MOLECULAR SCIENCE•NCE /W\/Q^DIRI DIRECT® PHYLOGENETICS AND EVOLUTION ELSEVIER Molecular Phylogenetics and Evolution 35 (2005) 147-164 www.elsevier.com/locate/ympev A multi-gene phylogeny of aquiline eagles (Aves: Accipitriformes) reveals extensive paraphyly at the genus level Andreas J. Helbig'^*, Annett Kocum'^, Ingrid Seibold^, Michael J. Braun^ '^ Institute of Zoology, University of Greifswald, Vogelwarte Hiddensee, D-18565 Kloster, Germany Department of Zoology, National Museum of Natural History, Smithsonian Institution, 4210 Silver Hill Rd., Suitland, MD 20746, USA Received 19 March 2004; revised 21 September 2004 Available online 24 December 2004 Abstract The phylogeny of the tribe Aquilini (eagles with fully feathered tarsi) was investigated using 4.2 kb of DNA sequence of one mito- chondrial (cyt b) and three nuclear loci (RAG-1 coding region, LDH intron 3, and adenylate-kinase intron 5). Phylogenetic signal was highly congruent and complementary between mtDNA and nuclear genes. In addition to single-nucleotide variation, shared deletions in nuclear introns supported one basal and two peripheral clades within the Aquilini. Monophyly of the Aquilini relative to other birds of prey was confirmed. However, all polytypic genera within the tribe, Spizaetus, Aquila, Hieraaetus, turned out to be non-monophyletic. Old World Spizaetus and Stephanoaetus together appear to be the sister group of the rest of the Aquilini. Spiza- stur melanoleucus and Oroaetus isidori axe nested among the New World Spizaetus species and should be merged with that genus. The Old World 'Spizaetus' species should be assigned to the genus Nisaetus (Hodgson, 1836). The sister species of the two spotted eagles (Aquila clanga and Aquila pomarina) is the African Long-crested Eagle (Lophaetus occipitalis). -

Application for Non-Resident Raptor Trapping Permit
Application for Non-Resident Raptor Trapping Permit Please complete this form and return it, along with the permit fee and requested attachments, to: Texas Parks and Wildlife Department, Wildlife Diversity Permits, 4200 Smith School Road, Austin, TX 78744. All applicants for a Non-Resident Raptor Trapping Permit must possess a permit from their home state equivalent to a Texas Apprentice, General or Master Falconer permit. A non-resident shall not st th trap more than one raptor per year in this state (July 1 – June 30 ). Applicant Name: Home Phone: Office Phone: Street Address: Cell Phone: Driver's License No.: City: State: Zip: Date of Birth: 1 Social Security E-mail Address: Number: 1. This application is for a Non-Resident Raptor Trapping Permit to allow a permitted non-resident apprentice, general or master falconer to take, from the wild in Texas with a valid non-resident hunting license, one of the following species of raptors within the permit year (July 1st – June 30th): Sharp-shinned hawk (Accipiter striatus), Cooper’s hawk (Accipiter cooperii), Harris’s Hawk (Parabuteo unicinctus), Red-shouldered hawk (Buteo lineatus), Red-tailed hawk (Buteo jamaicensis), Ferruginous hawk (Buteo regalis), American kestrel (Falco sparverius), Merlin (Falco columbarius), Prairie falcon (Falco mexicanus), Gyrfalcon (Falco rusticolus), or Caracara (Caracara cheriway). 2. Please attach a copy of your current state falconry permit. 3. Please enclose the $378.00 application and permit fee. (Make checks or money order payable to Texas Parks and Wildlife Department) The Texas Administrative Code Raptor Proclamation can be found online at: http://texreg.sos.state.tx.us/public/readtac$ext.ViewTAC?tac_view=5&ti=31&pt=2&ch=65&sch=K&rl=Y I have read and understand the state regulations titled “Raptor Proclamation” and I will comply with all state and federal laws pertaining to falconry. -

Chromosome Painting in Three Species of Buteoninae: a Cytogenetic Signature Reinforces the Monophyly of South American Species
Chromosome Painting in Three Species of Buteoninae: A Cytogenetic Signature Reinforces the Monophyly of South American Species Edivaldo Herculano C. de Oliveira1,2,3*, Marcella Mergulha˜o Tagliarini4, Michelly S. dos Santos5, Patricia C. M. O’Brien3, Malcolm A. Ferguson-Smith3 1 Laborato´rio de Cultura de Tecidos e Citogene´tica, SAMAM, Instituto Evandro Chagas, Ananindeua, PA, Brazil, 2 Faculdade de Cieˆncias Exatas e Naturais, ICEN, Universidade Federal do Para´, Bele´m, PA, Brazil, 3 Cambridge Resource Centre for Comparative Genomics, Cambridge, United Kingdom, 4 Programa de Po´s Graduac¸a˜oem Neurocieˆncias e Biologia Celular, ICB, Universidade Federal do Para´, Bele´m, PA, Brazil, 5 PIBIC – Universidade Federal do Para´, Bele´m, PA, Brazil Abstract Buteoninae (Falconiformes, Accipitridae) consist of the widely distributed genus Buteo, and several closely related species in a group called ‘‘sub-buteonine hawks’’, such as Buteogallus, Parabuteo, Asturina, Leucopternis and Busarellus, with unsolved phylogenetic relationships. Diploid number ranges between 2n = 66 and 2n = 68. Only one species, L. albicollis had its karyotype analyzed by molecular cytogenetics. The aim of this study was to present chromosomal analysis of three species of Buteoninae: Rupornis magnirostris, Asturina nitida and Buteogallus meridionallis using fluorescence in situ hybridization (FISH) experiments with telomeric and rDNA probes, as well as whole chromosome probes derived from Gallus gallus and Leucopternis albicollis. The three species analyzed herein showed similar karyotypes, with 2n = 68. Telomeric probes showed some interstitial telomeric sequences, which could be resulted by fusion processes occurred in the chromosomal evolution of the group, including the one found in the tassociation GGA1p/GGA6. -

Birds of the East Texas Baptist University Campus with Birds Observed Off-Campus During BIOL3400 Field Course
Birds of the East Texas Baptist University Campus with birds observed off-campus during BIOL3400 Field course Photo Credit: Talton Cooper Species Descriptions and Photos by students of BIOL3400 Edited by Troy A. Ladine Photo Credit: Kenneth Anding Links to Tables, Figures, and Species accounts for birds observed during May-term course or winter bird counts. Figure 1. Location of Environmental Studies Area Table. 1. Number of species and number of days observing birds during the field course from 2005 to 2016 and annual statistics. Table 2. Compilation of species observed during May 2005 - 2016 on campus and off-campus. Table 3. Number of days, by year, species have been observed on the campus of ETBU. Table 4. Number of days, by year, species have been observed during the off-campus trips. Table 5. Number of days, by year, species have been observed during a winter count of birds on the Environmental Studies Area of ETBU. Table 6. Species observed from 1 September to 1 October 2009 on the Environmental Studies Area of ETBU. Alphabetical Listing of Birds with authors of accounts and photographers . A Acadian Flycatcher B Anhinga B Belted Kingfisher Alder Flycatcher Bald Eagle Travis W. Sammons American Bittern Shane Kelehan Bewick's Wren Lynlea Hansen Rusty Collier Black Phoebe American Coot Leslie Fletcher Black-throated Blue Warbler Jordan Bartlett Jovana Nieto Jacob Stone American Crow Baltimore Oriole Black Vulture Zane Gruznina Pete Fitzsimmons Jeremy Alexander Darius Roberts George Plumlee Blair Brown Rachel Hastie Janae Wineland Brent Lewis American Goldfinch Barn Swallow Keely Schlabs Kathleen Santanello Katy Gifford Black-and-white Warbler Matthew Armendarez Jordan Brewer Sheridan A. -

Field Identification of the Field Identification of the Field
TOPICS IN IDENTIFICATION he Solitary Eagle ( Harpyhaliaetus solitarius ) is a large raptor that is closely related and similar in adult and immature plum- Tages to the black-hawks in the genus Buteogallus (Lerner and Mindell 2005). It is a rare and very local resident in a variety of wet and dry forested hills and highlands from northern Argentina to northern Mexico (del Hoyo et al. 1994, Ferguson-Lees and Christie 2001). The species has been collected in Mexico not far from the Texas border (see Discussion, pp. 72 –73), so it is possible that it has occurred in the ABA Area. The handful of specimens and nest records of this eagle are from 700 to 2,000 meters above sea level (Brown and Amadon 1968). FFiieelldd IIddeennttiifificcaattiioonn ooff tthhee SSOOLLIITTTAAARRRYYY EEAAAGGGLLLEEE Nevertheless, sightings of this eagle are occasionally reported from lowland tropical rain forest, e.g., at Tikal, Guatemala (Beaver et al. 1991) and the Tuxtlas Mountains of south - William S. Clark ern Veracruz, Mexico (Winker et al. 1992). The species has been reported on some pro - 2301 South Whitehouse Circle fessional bird tours at such lowland sites as Palenque and the Usumicinta River in south - Harlingen, Texas 78550 ern Mexico. All of these accounts have relied on large size and gray coloration as the [email protected] field marks to distinguish the eagles from the much more abundant Common Black- Hawk ( Buteogallus anthracinus ) and Great Black-Hawk ( B. urubitinga ). H. Lee Jones Howell and Webb (1995) were skeptical and stated that most lowland records of the 4810 Park Newport, No. -

Systematics and Conservation of the Hook-Billed Kite Including the Island Taxa from Cuba and Grenada J
Animal Conservation. Print ISSN 1367-9430 Systematics and conservation of the hook-billed kite including the island taxa from Cuba and Grenada J. A. Johnson1,2, R. Thorstrom1 & D. P. Mindell2 1 The Peregrine Fund, Boise, ID, USA 2 Department of Ecology & Evolutionary Biology, University of Michigan Museum of Zoology, Ann Arbor, MI, USA Keywords Abstract phylogenetics; coalescence; divergence; conservation; cryptic species; Chondrohierax. Taxonomic uncertainties within the genus Chondrohierax stem from the high degree of variation in bill size and plumage coloration throughout the geographic Correspondence range of the single recognized species, hook-billed kite Chondrohierax uncinatus. Jeff A. Johnson, Department of Ecology & These uncertainties impede conservation efforts as local populations have declined Evolutionary Biology, University of Michigan throughout much of its geographic range from the Neotropics in Central America Museum of Zoology, 1109 Geddes Avenue, to northern Argentina and Paraguay, including two island populations on Cuba Ann Arbor, MI 48109, USA. and Grenada, and it is not known whether barriers to dispersal exist between any Email: [email protected] of these areas. Here, we present mitochondrial DNA (mtDNA; cytochrome B and NADH dehydrogenase subunit 2) phylogenetic analyses of Chondrohierax, with Received 2 December 2006; accepted particular emphasis on the two island taxa (from Cuba, Chondrohierax uncinatus 20 April 2007 wilsonii and from Grenada, Chondrohierax uncinatus mirus). The mtDNA phylo- genetic results suggest that hook-billed kites on both islands are unique; however, doi:10.1111/j.1469-1795.2007.00118.x the Cuban kite has much greater divergence estimates (1.8–2.0% corrected sequence divergence) when compared with the mainland populations than does the Grenada hook-billed kite (0.1–0.3%). -

Bald Eagle Haliaeetus Leucocephalus
Wyoming Species Account Bald Eagle Haliaeetus leucocephalus REGULATORY STATUS USFWS: Delisted; Migratory Bird USFS R2: Sensitive USFS R4: Sensitive Wyoming BLM: Sensitive State of Wyoming: Protected Bird CONSERVATION RANKS USFWS: Bird of Conservation Concern WGFD: NSS3 (Bb), Tier II WYNDD: G5, S4B/S5N Wyoming Contribution: LOW IUCN: Least Concern PIF Continental Concern Score: 9 STATUS AND RANK COMMENTS Bald Eagle (Haliaeetus leucocephalus) is provided international protection under the Federal Migratory Bird Treaty Act of 1918, as amended 1. In 1940, Bald Eagle was provided protection under the Bald and Golden Eagle Protection Act 2. In 1966, the southern subspecies was listed as federally endangered under the Endangered Species Preservation Act; the entire population in the contiguous United States was listed as endangered in 1978 under the 1973 Endangered Species Act (ESA). A significant increase in numbers of nesting pairs, productivity, and distribution allowed Bald Eagle to be reclassified from Endangered to Threatened in 1995 under the ESA 3. Bald Eagle was delisted in 2007, and numbers are considered to be stable to increasing across its range 4. The species has been assigned different state conservation ranks by the Wyoming Natural Diversity Database for the breeding season and nonbreeding season because the abundance of the species is different between seasons. NATURAL HISTORY Taxonomy: Bald Eagle is a member of the family Accipitridae, which includes kites, eagles, harriers, and hawks 5. There are two subspecies of Bald Eagle; H. l. alascanus is found north of 40 degrees latitude across North America, including Wyoming, while H. l. leucocephalus is found south of 40 degrees latitude in the Gulf coast states 6. -

Regional Specialties Western
REGIONAL SPECIALTIES WESTERN OSPREY 21 - 26” length SOUTHERN . FERRUGINOUS . Eagle sized; clean, white body. HAWK Black wrist marks. 20 - 26” length . Glides with kink (M) in long, narrow wings. MISSISSIPPI . Largest buteo; eagle-like. KITE . Pale below with dark leggings. 13 - 15” length . Mostly white tail; 3 color morphs. Long, pointed wings; slim body. Light body; dark wings; narrow, black tail. Not to scale. Buoyant, acrobatic flight. NORTHERN HARRIER 16 - 20” length PRAIRIE FALCON 14 - 18” length . Long, narrow wings and tail; sharp dihedral. Size of Peregrine; much paler plumage. Brown above, streaked brown below – female. Narrow moustache; spotted breast; long tail. Gray above, pale below with black wing tips – male. Dark armpits and partial wing linings. WING PROFILE IMMATURE BALD EAGLE BALD EAGLE GOLDEN EAGLE . Immature birds vary GOLDEN EAGLE greatly in the amount 27 to 35” length of white spotting on body and wings. White showing on wing linings is surely a Bald Eagle. BALD EAGLE . Like large buteo, curvy wings. Head protrudes much less than tail. Slight dihedral to wing profile. NOTE: Some hawks soar and glide with their wings raised above the horizontal, called a dihedral. 27 to 35” length . Head and tail length similar. Long, flat wings. Straight leading edge to wings. 24 to 28” length This guide developed by Paul Carrier is the property of the Hawk Migration Association of North America (HMANA). HMANA is TURKey VUltUre a membership-based, non-profit organization committed to the . Dark wing linings with light flight feathers. conservation of raptors through the scientific study, enjoyment, and . Small head; long tail; sharp dihedral. -

Hawks & Falcons
[}{]&W[\~ ~ ~&[L©@ ~ ~ --- VERY SPECIAL BIRDS By Jer ry D. McG owan Game Biologist Fai rb an ks HAWKS ARE BIRDS OF PRE Y an d like other pred at o rs have long been persecuted be cau se t h ey fee ,, on other .',:" animals. But predators play an important ro le in the scheme of natu re and research is now begin nin g to reveal th e relat ion shi p of the se very specialized birds to t he environment . Most hawks fall int o o ne of th e three major groups t hat a year-round resident of Inte rior Alaska th at is most can be easily iden t ified wh en a bird is in flight. The buteos commonly fo un d in birch and aspen wo od s thro ughout the are soaring hawks wit h chu nky bo dies, b ro ad round ed state. wings, and fan -shaped tails . But eos found in Alaska are : Two other hawks nest in Alaska whic h are not membe rs the rough-legged hawk (Bute o lagopus), the red-tailed h awk of thes e three groups. They are the h arrier , or marsh hawk (Bu teo jamaic ensis] , -the Swainso n' s hawk (Buteo (Circus cyan eus) , and the ospre y (Pandion haliaetus) . The swainsoni) , the Harlans's hawk (Bueto harlan i) , the gold en marsh hawk is a slim bird with a white patch at the base eagle (Aquila chrysaeto s) , and th e bald eagle (Haliaeetus of the t ail, often seen flyi ng low with deliber ate wing b eat s. -
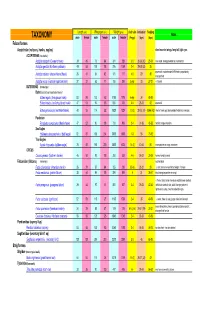
Avian Taxonomy
Length (cm) Wing span (cm) Weight (gms) cluch size incubation fledging Notes TAXONOMY male female male female male female (# eggs) (days) (days) Falconiformes Accipitridae (vultures, hawks, eagles) short rounded wings; long tail; light eyes ACCIPITRINAE (true hawks) Accipiter cooperii (Cooper's hawk) 39 45 73 84 341 528 3-5 30-36 (30) 25-34 crow sized; strongly banded tail; rounded tail Accipiter gentilis (Northern goshawk) 49 58 101 108 816 1059 2-4 28-38 (33) 35 square tail; most abundant NAM hawk; proportionaly 26 31 54 62 101 177 4-5 29 30 Accipiter striatus (sharp-shinned hawk) strongest foot Accipiter nisus (Eurasian sparrowhawk ) 37 37 62 77 150 290 5-Apr 33 27-31 Musket BUTEONINAE (broadwings) Buteo (buzzards or broad tailed hawks) Buteo regalis (ferruginous hawk) 53 59 132 143 1180 1578 6-Apr 34 45-50 Buteo lineatus (red-shouldered hawk) 47 53 96 105 550 700 3-4 28-33 42 square tail Buteo jamaicensis (red-tailed hawk) 48 55 114 122 1028 1224 1-3 (3) 28-35 (34) 42-46 (42) (Harlan' hawk spp); dark patagial featehres: immature; Parabuteo Parabuteo cuncincutus (Harris hawk) 47 52 90 108 710 890 2-4 31-36 45-50 reddish orange shoulders Sea Eagles Haliaeetus leucocephalus (bald eagle) 82 87 185 244 3000 6300 1-3 35 70-92 True Eagles Aquila chrysaetos (golden eagle) 78 82 185 220 3000 6125 1-4 (2) 40-45 50 white patches on wings: immature; CIRCUS Circus cyaneus (Northern harrier) 46 50 93 108 350 530 4-6 26-32 30-35 hovers; hunts by sound Falconidae (falcons) (longwings) notched beak Falco columbarius (American merlin) 26 29 57 64